In Vivo Microbiome and Associated Immune Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lactobacillus Crispatus
WHAT 2nd Microbiome workshop WHEN 17 & 18 November 2016 WHERE Masur Auditorium at NIH Campus, Bethesda MD Modulating the Vaginal Microbiome to Prevent HIV Infection Laurel Lagenaur For more information: www.virology-education.com Conflict of Interest I work for Osel Inc., a microbiome company developing live biotherapeutic products to prevent diseases in women The Vaginal Microbiome and HIV Infection What’s normal /healthy/ optimal and what’s not? • Ravel Community Groups • Lactobacillus dominant vs. dysbiosis Why are Lactobacilli important for HIV prevention? • Dysbiosis = Inflammation = Increased risk of HIV acquisition • Efficacy of Pre-Exposure Prophylaxis decreased Modulation of the vaginal microbiome • LACTIN-V, live biotherapeutic-Lactobacillus crispatus • Ongoing clinical trials How we can use Lactobacilli and go a step further • Genetically modified Lactobacillus to prevent HIV acquisition HIV is Transmitted Across Mucosal Surfaces • HIV infection in women occurs in the mucosa of the vagina and cervix vagina cervix • Infection of underlying target cells All mucosal surfaces are continuously exposed to a Herrera and Shattock, community of microorganisms Curr Top Microbiol Immunol 2013 Vaginal Microbiome: Ravel Community Groups Vaginal microbiomes clustered into 5 groups: Group V L. jensenii Group II L. gasseri 4 were dominated by Group I L. crispatus Lactobacillus, Group III L. iners whereas the 5th had lower proportions of lactic acid bacteria and Group 4 Diversity higher proportions of Prevotella, strictly anaerobic Sneathia, -

A Taxonomic Note on the Genus Lactobacillus
Taxonomic Description template 1 A taxonomic note on the genus Lactobacillus: 2 Description of 23 novel genera, emended description 3 of the genus Lactobacillus Beijerinck 1901, and union 4 of Lactobacillaceae and Leuconostocaceae 5 Jinshui Zheng1, $, Stijn Wittouck2, $, Elisa Salvetti3, $, Charles M.A.P. Franz4, Hugh M.B. Harris5, Paola 6 Mattarelli6, Paul W. O’Toole5, Bruno Pot7, Peter Vandamme8, Jens Walter9, 10, Koichi Watanabe11, 12, 7 Sander Wuyts2, Giovanna E. Felis3, #*, Michael G. Gänzle9, 13#*, Sarah Lebeer2 # 8 '© [Jinshui Zheng, Stijn Wittouck, Elisa Salvetti, Charles M.A.P. Franz, Hugh M.B. Harris, Paola 9 Mattarelli, Paul W. O’Toole, Bruno Pot, Peter Vandamme, Jens Walter, Koichi Watanabe, Sander 10 Wuyts, Giovanna E. Felis, Michael G. Gänzle, Sarah Lebeer]. 11 The definitive peer reviewed, edited version of this article is published in International Journal of 12 Systematic and Evolutionary Microbiology, https://doi.org/10.1099/ijsem.0.004107 13 1Huazhong Agricultural University, State Key Laboratory of Agricultural Microbiology, Hubei Key 14 Laboratory of Agricultural Bioinformatics, Wuhan, Hubei, P.R. China. 15 2Research Group Environmental Ecology and Applied Microbiology, Department of Bioscience 16 Engineering, University of Antwerp, Antwerp, Belgium 17 3 Dept. of Biotechnology, University of Verona, Verona, Italy 18 4 Max Rubner‐Institut, Department of Microbiology and Biotechnology, Kiel, Germany 19 5 School of Microbiology & APC Microbiome Ireland, University College Cork, Co. Cork, Ireland 20 6 University of Bologna, Dept. of Agricultural and Food Sciences, Bologna, Italy 21 7 Research Group of Industrial Microbiology and Food Biotechnology (IMDO), Vrije Universiteit 22 Brussel, Brussels, Belgium 23 8 Laboratory of Microbiology, Department of Biochemistry and Microbiology, Ghent University, Ghent, 24 Belgium 25 9 Department of Agricultural, Food & Nutritional Science, University of Alberta, Edmonton, Canada 26 10 Department of Biological Sciences, University of Alberta, Edmonton, Canada 27 11 National Taiwan University, Dept. -

Interstrain Variability of Human Vaginal Lactobacillus Crispatus for Metabolism of Biogenic Amines and Antimicrobial Activity Against Urogenital Pathogens
molecules Article Interstrain Variability of Human Vaginal Lactobacillus crispatus for Metabolism of Biogenic Amines and Antimicrobial Activity against Urogenital Pathogens Scarlett Puebla-Barragan 1,2 , Emiley Watson 1,2, Charlotte van der Veer 3 , John A. Chmiel 1,2 , Charles Carr 1, Jeremy P. Burton 1,2 , Mark Sumarah 4 , Remco Kort 5,6 and Gregor Reid 1,2,* 1 Canadian Centre for Human Microbiome and Probiotics, Lawson Health Research Institute, 268 Grosvenor Street, London, ON N6A 4V2, Canada; [email protected] (S.P.-B.); [email protected] (E.W.); [email protected] (J.A.C.); [email protected] (C.C.); [email protected] (J.P.B.) 2 Departments of Microbiology and Immunology, and Surgery, Western University, London, ON N6A 4V2, Canada 3 Department of Infectious Diseases, Public Health Service (GGD), Nieuwe Achtergracht 100, 1018 WT Amsterdam, The Netherlands; [email protected] 4 Agriculture and Agri-Food Canada, London, ON N5V 4T3, Canada; [email protected] 5 Department of Molecular Cell Biology, Faculty of Science, O2 Lab Building, Vrije Universiteit Amsterdam, De Boelelaan 1108, 1081 HZ Amsterdam, The Netherlands; [email protected] 6 ARTIS-Micropia, Plantage Kerklaan 38-40, 1018 CZ Amsterdam, The Netherlands Citation: Puebla-Barragan, S.; * Correspondence: [email protected]; Tel.: +1-519-646-6100 (ext. 65256) Watson, E.; van der Veer, C.; Chmiel, J.A.; Carr, C.; Burton, J.P.; Sumarah, Abstract: Lactobacillus crispatus is the dominant species in the vagina of many women. With the M.; Kort, R.; Reid, G. Interstrain potential for strains of this species to be used as a probiotic to help prevent and treat dysbiosis, we Variability of Human Vaginal investigated isolates from vaginal swabs with Lactobacillus-dominated and a dysbiotic microbiota. -

BD™ Gardnerella Selective Agar with 5% Human Blood
INSTRUCTIONS FOR USE – READY-TO-USE PLATED MEDIA PA-254094.06 Rev.: July 2014 BD Gardnerella Selective Agar with 5% Human Blood INTENDED USE BD Gardnerella Selective Agar with 5% Human Blood is a partially selective and differential medium for the isolation of Gardnerella vaginalis from clinical specimens. PRINCIPLES AND EXPLANATION OF THE PROCEDURE Microbiological method. Gardnerella vaginalis is considered to be one of the organisms causing vaginitis.1-4 Although the organism may be present in a high percentage of normal women in the vaginal flora, its importance as a cause of non-specific vaginitis (also called bacterial vaginosis) has never been questioned. In symptomatic women, G. vaginalis frequently is associated with anaerobes such as Prevotella bivia, P. disiens, Mobiluncus, Peptostreptococcus, and/or others which are a regular part of the urethral or intestinal, but not vaginal flora. In non-specific vaginitis, normal Lactobacillus flora is reduced or absent. Gardnerella vaginalis is considered to be the indicator organism for non-specific vaginitis which, in fact, is a polymicrobial infection.3,4 Although non- culture methods such as a direct Gram stain have been recommended in recent years for genital specimens, culture is still preferred by many laboratories.1,5 G. vaginalis may also be responsible for a variety of other diseases such as preterm birth, chorioamnionitis, urinary tract infections, newborn infections, and septicemia.6 The detection of the organism on routinely used media is difficult since Gardnerella and other -

Vaginal Probiotic Lactobacillus Crispatus Seems to Inhibit Sperm Activity and Subsequently Reduces Pregnancies in Rat
fcell-09-705690 August 11, 2021 Time: 11:32 # 1 ORIGINAL RESEARCH published: 13 August 2021 doi: 10.3389/fcell.2021.705690 Vaginal Probiotic Lactobacillus crispatus Seems to Inhibit Sperm Activity and Subsequently Reduces Pregnancies in Rat Ping Li1, Kehong Wei1, Xia He2, Lu Zhang1, Zhaoxia Liu3, Jing Wei1, Xiaomei Chen1, Hong Wei4* and Tingtao Chen1* 1 School of Life Sciences, Institute of Translational Medicine, Nanchang University, Nanchang, China, 2 Department of Obstetrics and Gynecology, The Ninth Hospital of Nanchang, Nanchang, China, 3 Department of Obstetrics and Gynecology, The Second Affiliated Hospital of Nanchang University, Nanchang, China, 4 Institute of Precision Medicine, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China Background: The vaginal microbiota is associated with the health of the female reproductive system and the offspring. Lactobacillus crispatus belongs to one of the most important vaginal probiotics, while its role in the agglutination and immobilization Edited by: of human sperm, fertility, and offspring health is unclear. Bechan Sharma, University of Allahabad, India Methods: Adherence assays, sperm motility assays, and Ca2C-detecting assays were Reviewed by: used to analyze the adherence properties and sperm motility of L. crispatus Lcr-MH175, António Machado, Universidad San Francisco de Quito, attenuated Salmonella typhimurium VNP20009, engineered S. typhimurium VNP20009 Ecuador DNase I, and Escherichia coli O157:H7 in vitro. The rat reproductive model was further Margarita Aguilera, University of Granada, Spain developed to study the role of L. crispatus on reproduction and offspring health, using *Correspondence: high-throughput sequencing, real-time PCR, and molecular biology techniques. Tingtao Chen Our results indicated that L. -

Human Microbiota Network: Unveiling Potential Crosstalk Between the Different Microbiota Ecosystems and Their Role in Health and Disease
nutrients Review Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease Jose E. Martínez †, Augusto Vargas † , Tania Pérez-Sánchez , Ignacio J. Encío , Miriam Cabello-Olmo * and Miguel Barajas * Biochemistry Area, Department of Health Science, Public University of Navarre, 31008 Pamplona, Spain; [email protected] (J.E.M.); [email protected] (A.V.); [email protected] (T.P.-S.); [email protected] (I.J.E.) * Correspondence: [email protected] (M.C.-O.); [email protected] (M.B.) † These authors contributed equally to this work. Abstract: The human body is host to a large number of microorganisms which conform the human microbiota, that is known to play an important role in health and disease. Although most of the microorganisms that coexist with us are located in the gut, microbial cells present in other locations (like skin, respiratory tract, genitourinary tract, and the vaginal zone in women) also play a significant role regulating host health. The fact that there are different kinds of microbiota in different body areas does not mean they are independent. It is plausible that connection exist, and different studies have shown that the microbiota present in different zones of the human body has the capability of communicating through secondary metabolites. In this sense, dysbiosis in one body compartment Citation: Martínez, J.E.; Vargas, A.; may negatively affect distal areas and contribute to the development of diseases. Accordingly, it Pérez-Sánchez, T.; Encío, I.J.; could be hypothesized that the whole set of microbial cells that inhabit the human body form a Cabello-Olmo, M.; Barajas, M. -

The Microbiota Continuum Along the Female Reproductive Tract and Its Relation to Uterine-Related Diseases
ARTICLE DOI: 10.1038/s41467-017-00901-0 OPEN The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases Chen Chen1,2, Xiaolei Song1,3, Weixia Wei4,5, Huanzi Zhong 1,2,6, Juanjuan Dai4,5, Zhou Lan1, Fei Li1,2,3, Xinlei Yu1,2, Qiang Feng1,7, Zirong Wang1, Hailiang Xie1, Xiaomin Chen1, Chunwei Zeng1, Bo Wen1,2, Liping Zeng4,5, Hui Du4,5, Huiru Tang4,5, Changlu Xu1,8, Yan Xia1,3, Huihua Xia1,2,9, Huanming Yang1,10, Jian Wang1,10, Jun Wang1,11, Lise Madsen 1,6,12, Susanne Brix 13, Karsten Kristiansen1,6, Xun Xu1,2, Junhua Li 1,2,9,14, Ruifang Wu4,5 & Huijue Jia 1,2,9,11 Reports on bacteria detected in maternal fluids during pregnancy are typically associated with adverse consequences, and whether the female reproductive tract harbours distinct microbial communities beyond the vagina has been a matter of debate. Here we systematically sample the microbiota within the female reproductive tract in 110 women of reproductive age, and examine the nature of colonisation by 16S rRNA gene amplicon sequencing and cultivation. We find distinct microbial communities in cervical canal, uterus, fallopian tubes and perito- neal fluid, differing from that of the vagina. The results reflect a microbiota continuum along the female reproductive tract, indicative of a non-sterile environment. We also identify microbial taxa and potential functions that correlate with the menstrual cycle or are over- represented in subjects with adenomyosis or infertility due to endometriosis. The study provides insight into the nature of the vagino-uterine microbiome, and suggests that sur- veying the vaginal or cervical microbiota might be useful for detection of common diseases in the upper reproductive tract. -

Lactobacillus Crispatus Protects Against Bacterial Vaginosis
Lactobacillus crispatus protects against bacterial vaginosis M.O. Almeida1, F.L.R. do Carmo1, A. Gala-García1, R. Kato1, A.C. Gomide1, R.M.N. Drummond2, M.M. Drumond3, P.M. Agresti1, D. Barh4, B. Brening5, P. Ghosh6, A. Silva7, V. Azevedo1 and 1,7 M.V.C. Viana 1 Departamento de Genética, Ecologia e Evolução, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brasil 2 Departamento de Microbiologia, Ecologia e Evolução, , Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brasil 3 Departamento de Ciências Biológicas, Centro Federal de Educação Tecnologica de Minas Gerais, Belo Horizonte, MG, Brasil 4 Institute of Integrative Omics and Applied Biotechnology (IIOAB), Nonakuri, Purba Medinipur, West Bengal, India 5 Institute of Veterinary Medicine, University of Göttingen, Göttingen, Germany 6 Department of Computer Science, Virginia Commonwealth University, Richmond, Virginia, USA 7 Departamento de Genética, Instituto de Ciências Biológicas, Universidade Federal do Pará, Belém, PA, Brasil Corresponding author: V. Azevedo E-mail: [email protected] Genet. Mol. Res. 18 (4): gmr18475 Received August 16, 2019 Accepted October 23, 2019 Published November 30, 2019 DOI http://dx.doi.org/10.4238/gmr18475 ABSTRACT. In medicine, the 20th century was marked by one of the most important revolutions in infectious-disease management, the discovery and increasing use of antibiotics. However, their indiscriminate use has led to the emergence of multidrug-resistant (MDR) bacteria. Drug resistance and other factors, such as the production of bacterial biofilms, have resulted in high recurrence rates of bacterial diseases. Bacterial vaginosis (BV) syndrome is the most prevalent vaginal condition in women of reproductive age, Genetics and Molecular Research 18 (4): gmr18475 ©FUNPEC-RP www.funpecrp.com.br M.O. -
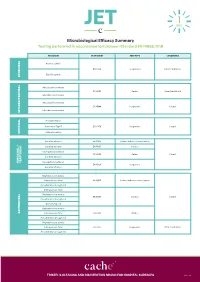
JET Microbiological Efficacy Summary
Microbiological Efficacy Summary Testing performed in accordance to European Standard EN 14885:2018 ORGANISM TEST NORM TEST TYPE CONDITIONS Bacillus subtilis EN 13704 Suspension Clean 1 and Dirty 1 Bacillus cereus SPORICIDAL Mycobacterium terrae EN 14563 Carrier Clean 1 and Dirty 2 Mycobacterium avium Mycobacterium terrae EN 14348 Suspension Clean 1 Mycobacterium avium MYCOBACTERICIDAL Poliovirus Type 1 Adenovirus Type 5 EN 14476 Suspension Clean 1 Murine Norovirus VIRUCIDAL Candida albicans EN 16615 Surface with mechanical action Candida albicans EN 13697 Surface Aspergillus brasiliensis EN 14562 Carrier Clean 1 Candida albicans YEASTICIDAL FUNGICIDAL / FUNGICIDAL Aspergillus brasiliensis EN 13624 Suspension Candida albicans Staphylococcus aureus Enterococcus hirae EN 16615 Surface with mechanical action Pseudomonas aeruginosa Enterococcus hirae Staphylococcus aureus EN 13697 Surface Clean 1 Pseudomonas aeruginosa Escherichia coli Staphylococcus aureus BACTERICIDAL Enterococcus hirae EN 14561 Carrier Pseudomonas aeruginosa Staphylococcus aureus Enterococcus hirae EN 13727 Suspension Clean 1 and Dirty 1 Pseudomonas aeruginosa TRISTEL’S CLEANING AND DISINFECTION BRAND FOR HOSPITAL SURFACES Page 1 of 3 Additional Testing TEST METHOD RNA DNA / Polyacrylamide gel electrophoresis (PAGE) ORGANISM TEST METHOD TEST TYPE CONDITIONS Acanthamoeba castellanii cysts Following the method of EN 13704 Suspension Clean 1 PROTOZOA Bacillus subtilis EN 17126 Suspension Clean 1 Bacillus cereus Clostridium difficile EN 13704 Suspension Clean 1 and Dirty 1 -

NOTES in Vitro Activities of Norfloxacin and Ciprofloxacin Against
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 1984, p. 94-96 Vol. 26, No. 1 0066-4804/84/070094-03$02.00/0 Copyright C 1984, American Society for Microbiology NOTES In Vitro Activities of Norfloxacin and Ciprofloxacin Against Mycobacterium tuberculosis, M. avium Complex, M. chelonei, M. fortuitum, and M. kansasii J. DOUGLAS GAY, DONALD R. DEYOUNG, AND GLENN D. ROBERTS* Section of Clinical Microbiology, Department of Laboratory Medicine, Mayo Clinic and Mayo Foundation, Rochester, Minnesota 55905 Received 28 November 1983/Accepted 4 April 1984 The activities of ciprofloxacin and norfloxacin against 100 mycobacteria isolates were studied in vitro by the 1% standard proportion method. Ciprofloxacin was more active against M. tuberculosis and M. fortuitum with MICs of 1.0 and 0.25 ,ug/ml, respectively, against 90% of isolates; norfloxacin had MICs of 8.0 and 2.0 ,ug/ml, respectively, against 90% of isolates. Nalidixic acid and other heterocyclic carbonic acid deriva- studied. The organisms were taken from the Mayo Clinic tives have been used primarily in the treatment of urinary stock culture collection, which included recent clinical iso- tract infections for many years. The compounds of this lates. Stock cultures were maintained on Middlebrook 7H10 general group include nalidixic acid, oxolinic acid, pipemidic agar slants (Difco Laboratories, Detroit, Mich.) and were acid, cinoxacin, and rosoxacin. Two new substances in this subcultured monthly. The identification of isolates was series which have been recently synthesized are norfloxacin based on standard biochemical tests (17) and gas-liquid (6) (1-ethyl-6-fluoro-1,4-dihydro-4-oxo-7-[ 1-piperazinyl ]-3- chromatography (16). -

A Genomic Journey Through a Genus of Large DNA Viruses
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2013 Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses Adrien Jeanniard Aix-Marseille Université David D. Dunigan University of Nebraska-Lincoln, [email protected] James Gurnon University of Nebraska-Lincoln, [email protected] Irina V. Agarkova University of Nebraska-Lincoln, [email protected] Ming Kang University of Nebraska-Lincoln, [email protected] See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/virologypub Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cell and Developmental Biology Commons, Genetics and Genomics Commons, Infectious Disease Commons, Medical Immunology Commons, Medical Pathology Commons, and the Virology Commons Jeanniard, Adrien; Dunigan, David D.; Gurnon, James; Agarkova, Irina V.; Kang, Ming; Vitek, Jason; Duncan, Garry; McClung, O William; Larsen, Megan; Claverie, Jean-Michel; Van Etten, James L.; and Blanc, Guillaume, "Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses" (2013). Virology Papers. 245. https://digitalcommons.unl.edu/virologypub/245 This Article is brought to you for free and open access by the Virology, Nebraska Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Virology Papers by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Adrien Jeanniard, David D. Dunigan, James Gurnon, Irina V. Agarkova, Ming Kang, Jason Vitek, Garry Duncan, O William McClung, Megan Larsen, Jean-Michel Claverie, James L. Van Etten, and Guillaume Blanc This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ virologypub/245 Jeanniard, Dunigan, Gurnon, Agarkova, Kang, Vitek, Duncan, McClung, Larsen, Claverie, Van Etten & Blanc in BMC Genomics (2013) 14. -

A Taxonomic Note on the Genus Lactobacillus
TAXONOMIC DESCRIPTION Zheng et al., Int. J. Syst. Evol. Microbiol. DOI 10.1099/ijsem.0.004107 A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae Jinshui Zheng1†, Stijn Wittouck2†, Elisa Salvetti3†, Charles M.A.P. Franz4, Hugh M.B. Harris5, Paola Mattarelli6, Paul W. O’Toole5, Bruno Pot7, Peter Vandamme8, Jens Walter9,10, Koichi Watanabe11,12, Sander Wuyts2, Giovanna E. Felis3,*,†, Michael G. Gänzle9,13,*,† and Sarah Lebeer2† Abstract The genus Lactobacillus comprises 261 species (at March 2020) that are extremely diverse at phenotypic, ecological and gen- otypic levels. This study evaluated the taxonomy of Lactobacillaceae and Leuconostocaceae on the basis of whole genome sequences. Parameters that were evaluated included core genome phylogeny, (conserved) pairwise average amino acid identity, clade- specific signature genes, physiological criteria and the ecology of the organisms. Based on this polyphasic approach, we propose reclassification of the genus Lactobacillus into 25 genera including the emended genus Lactobacillus, which includes host- adapted organisms that have been referred to as the Lactobacillus delbrueckii group, Paralactobacillus and 23 novel genera for which the names Holzapfelia, Amylolactobacillus, Bombilactobacillus, Companilactobacillus, Lapidilactobacillus, Agrilactobacil- lus, Schleiferilactobacillus, Loigolactobacilus, Lacticaseibacillus, Latilactobacillus, Dellaglioa,