And Libythea Celtis (Nymphalidae: Libytheinae) and Their Phylogenetic Implications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Variation in Butterfly Diversity and Unique Species Richness Along
Check List 8(3): 432-436, 2012 © 2012 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution PECIES S OF Sanctuary,Variation in Tripura, butterfly northeast diversity India and unique species ISTS L richness along different habitats in Trishna Wildlife * Joydeb Majumder, Rahul Lodh and B. K. Agarwala [email protected] Tripura University, Department of Zoology, Ecology and Biodiversity Laboratories, Suryamaninagar – 799 022, Tripura, India. * Corresponding author. E-mail: Abstract: Quantification of butterfly diversity and species richness is of prime importance for evaluating the status of protected areas. Permanent line transect counts were used to record species richness and abundance of butterfly communities of different habitat types in Trishna wildlife sanctuary. A total of 1005 individuals representing 59 species in 48 genera belonging to five families were recorded in the present study. Of these, 23 species belonged to the family Nymphalidae and accounted for 38.98% of the total species and 45.20% of the total number of individuals. Mature secondary mixed moist deciduous forest showed the maximum diversity and species richness, while exotic grassland showed minimum diversity and species richness. Out of 59 species, 31 are new records for Tripura state, while 21 are unique species and nine are listed in the threatened category. This study revealed that mature secondary forests are more important for butterfly communities, while exotic grasslands have a negative impact on species composition. Introduction state of Tripura (10,490 sq km), in northeastern India In the humid tropics, due to deforestation of primary (Mandal et al. -

The Complete Mitochondrial Genome of the Fall Webworm, Hyphantria
Int. J. Biol. Sci. 2010, 6 172 International Journal of Biological Sciences 2010; 6(2):172-186 © Ivyspring International Publisher. All rights reserved Research Paper The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae) Fang Liao1,2, Lin Wang3, Song Wu4, Yu-Ping Li4, Lei Zhao1, Guo-Ming Huang2, Chun-Jing Niu2, Yan-Qun Liu4, , Ming-Gang Li1, 1. College of Life Sciences, Nankai University, Tianjin 300071, China 2. Tianjin Entry-Exit Inspection and Quarantine Bureau, Tianjin 300457, China 3. Beijing Entry-Exit Inspection and Quarantine Bureau, Beijing 101113, China 4. College of Bioscience and Biotechnology, Shenyang Agricultural University, Liaoning, Shenyang 110866, China Corresponding author: Y. Q. Liu, College of Bioscience and Biotechnology, Shenyang Agricultural University, Liaoning, Shenyang 110866, China; Tel: 86-24-88487163; E-mail: [email protected]. Or to: M. G. Li, College of Life Sciences, Nankai University, Tianjin 300071, China; Tel: 86-22-23508237; E-mail: [email protected]. Received: 2010.02.03; Accepted: 2010.03.26; Published: 2010.03.29 Abstract The complete mitochondrial genome (mitogenome) of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae) was determined. The genome is a circular molecule 15 481 bp long. It presents a typical gene organization and order for completely sequenced lepidopteran mitogenomes, but differs from the insect ancestral type for the placement of tRNAMet. The nucleotide composition of the genome is also highly A + T biased, accounting for 80.38%, with a slightly positive AT skewness (0.010), indicating the occurrence of more As than Ts, as found in the Noctuoidea species. All protein-coding genes (PCGs) are initiated by ATN codons, except for COI, which is tentatively designated by the CGA codon as observed in other le- pidopterans. -

Taxonomic Revision of the Tribe Danaini (Lepidoptera: Nymphalidae: Danainae) from Myanmar
JAPB191_proof ■ 5 February 2017 ■ 1/5 Journal of Asia-Pacific Biodiversity xxx (2017) 1e5 55 HOSTED BY Contents lists available at ScienceDirect 56 57 Journal of Asia-Pacific Biodiversity 58 59 60 journal homepage: http://www.elsevier.com/locate/japb 61 62 63 Original article 64 65 1 Taxonomic revision of the tribe Danaini (Lepidoptera: Nymphalidae: 66 2 67 3 Danainae) from Myanmar 68 4 69 a a a b a,* 5 Q4 Nan Zarchi Win , Eun Young Choi , Jong Bong Choi , Jinyoung Park , Jong Kyun Park 70 6 a 71 7 College of Ecology and Environmental Science, Kyungpook National University, Sangju, Republic of Korea b Department of Nature Survey, National Institute of Ecology, Seocheon, Republic of Korea 72 8 73 9 74 10 article info abstract 75 11 76 12 Article history: The tribe Danaini is reviewed for the first time from Myanmar. Ten species of four genera belonging to 77 13 Received 29 September 2016 two subtribes are taxonomically described. Identification keys for the subtribes, the genera, and all 78 14 Received in revised form species are provided. The adult illustrations for all examined species are also presented. 79 8 November 2016 15 Copyright Ó 2017, National Science Museum of Korea (NSMK) and Korea National Arboretum (KNA). 80 Accepted 11 November 2016 16 Production and hosting by Elsevier. This is an open access article under the CC BY-NC-ND license (http:// Available online xxx 81 17 creativecommons.org/licenses/by-nc-nd/4.0/). 82 18 Keywords: 83 19 butterfly 84 20 Danaini 85 Danainae 21 86 Myanmar 22 87 23 88 24 89 25 Introduction Myanmar is one of the biologically diverse countries in main- 90 26 land Southeast Asia and rich in biodiversity. -

The Complete Mitochondrial Genome of Hipparchia Autonoe (Esper, 1783) (Lepidoptera: Nymphalidae): Investigation of Intraspecific Variations on Mitochondrial Genome
Mitochondrial DNA Part B Resources ISSN: (Print) 2380-2359 (Online) Journal homepage: https://www.tandfonline.com/loi/tmdn20 The complete mitochondrial genome of Hipparchia autonoe (Esper, 1783) (Lepidoptera: Nymphalidae): investigation of intraspecific variations on mitochondrial genome Yeong-Don Lee, Jungmo Lee, Do-Sung Kim, Jonghyun Park, Hong Xi, Jeehee Roh, Dong-Soon Kim, Sang June Nam, Seong-Ki Kim, Jin-Young Song & Jongsun Park To cite this article: Yeong-Don Lee, Jungmo Lee, Do-Sung Kim, Jonghyun Park, Hong Xi, Jeehee Roh, Dong-Soon Kim, Sang June Nam, Seong-Ki Kim, Jin-Young Song & Jongsun Park (2020) The complete mitochondrial genome of Hipparchiaautonoe (Esper, 1783) (Lepidoptera: Nymphalidae): investigation of intraspecific variations on mitochondrial genome, Mitochondrial DNA Part B, 5:2, 1542-1544, DOI: 10.1080/23802359.2020.1742230 To link to this article: https://doi.org/10.1080/23802359.2020.1742230 © 2020 The Author(s). Published by Informa Published online: 24 Mar 2020. UK Limited, trading as Taylor & Francis Group. Submit your article to this journal Article views: 95 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at https://www.tandfonline.com/action/journalInformation?journalCode=tmdn20 MITOCHONDRIAL DNA PART B 2020, VOL. 5, NO. 2, 1542–1544 https://doi.org/10.1080/23802359.2020.1742230 MITOGENOME ANNOUNCEMENT The complete mitochondrial genome of Hipparchia autonoe (Esper, 1783) (Lepidoptera: Nymphalidae): investigation of intraspecific variations on mitochondrial -

Diversity of Takhni Rehmapur Wildlife Sanctuary, Hoshiarpur, Punjab, India
SHARMA : Butterfly Diversity of Takhni Rehmapur Wildlife Sanctuary, Hoshiarpur, Punjab, India 305 ISSN 0375-1511 Rec. zool. Surv. India : 115(Part-3) : 305-311, 2015 BUTTERFLY (LEPIDOPTERA: INSECTA) DIVERSITY OF TAKHNI REHMAPUR WILDLIFE SANCTUARY, HOSHIARPUR, PUNJAB, INDIA NARENDER SHARMA Zoological Survey of India, Northern Regional Centre, 218 Kaulagarh Road, Dehradun-248 195 Email : [email protected] INTRODUCTION the available information is mainly restricted to The butterfly fauna of India has been well that published by Rose and Sidhu (2001), who studied in the past, with the works of Marshall & de provided an inventory of 74 species of butterflies Niceville (1883), de Niceville (1886, 1890), Moore from Punjab; Arora et al. (2006), who gave a brief (1890-1905), Swinhoe (1893, 1896, 1905-1913), account of 74 species from Punajb Shivaliks; and Bingham (1905, 1907), Evans (1932), Talbot Sharma and Joshi (2009), who listed 41 species (1939, 1947), Wynter-Blyth (1957), and Kehimkar from Dholbaha Dam (Hoshiarpur). However, (2008) being some of the significant publications. information on the butterfly diversity of the various To date, 1641 species of butterflies have been protected areas of Punjab is almost totally lacking. reported from India (Varshney, 2010). Recently, It is precisely with this point in mind that while much information on butterflies of different conducting ‘General Faunistic Surveys’ of Punjab regions, states and protected areas of India has under the mandates of the Zoological Survey of been published (e.g. Arora et al. 2009 (Himachal India, we were fortunate to have the opportunity Pradesh); Anonymous (website of Punjab ENVIS to study the butterfly faunal diversity of Takhni Centre, Punjab); Kumar 2008 (Uttarakhand); Rehmapur Wildlife Sanctuary on 12th and 13th Mondal et al., 1997 (Delhi); Chandra et al. -

Development of Encyclopedia Boyong Sleman Insekta River As Alternative Learning Resources
PROC. INTERNAT. CONF. SCI. ENGIN. ISSN 2597-5250 Volume 3, April 2020 | Pages: 629-634 E-ISSN 2598-232X Development of Encyclopedia Boyong Sleman Insekta River as Alternative Learning Resources Rini Dita Fitriani*, Sulistiyawati Biological Education Faculty of Science and Technology, UIN Sunan Kalijaga Jl. Marsda Adisucipto Yogyakarta, Indonesia Email*: [email protected] Abstract. This study aims to determine the types of insects Coleoptera, Hemiptera, Odonata, Orthoptera and Lepidoptera in the Boyong River, Sleman Regency, Yogyakarta, to develop the Encyclopedia of the Boyong River Insect and to determine the quality of the encyclopedia developed. The method used in the research inventory of the types of insects Coleoptera, Hemiptera, Odonata, Orthoptera and Lepidoptera insects in the Boyong River survey method with the results of the study found 46 species of insects consisting of 2 Coleoptera Orders, 2 Hemiptera Orders, 18 orders of Lepidoptera in Boyong River survey method with the results of the research found 46 species of insects consisting of 2 Coleoptera Orders, 2 Hemiptera Orders, 18 orders of Lepidoptera in Boyong River survey method. odonata, 4 Orthopterous Orders and 20 Lepidopterous Orders from 15 families. The encyclopedia that was developed was created using the Adobe Indesig application which was developed in printed form. Testing the quality of the encyclopedia uses a checklist questionnaire and the results of the percentage of ideals from material experts are 91.1% with very good categories, 91.7% of media experts with very good categories, peer reviewers 92.27% with very good categories, biology teachers 88, 53% with a very good category and students 89.8% with a very good category. -

Evolution of Insect Color Vision: from Spectral Sensitivity to Visual Ecology
EN66CH23_vanderKooi ARjats.cls September 16, 2020 15:11 Annual Review of Entomology Evolution of Insect Color Vision: From Spectral Sensitivity to Visual Ecology Casper J. van der Kooi,1 Doekele G. Stavenga,1 Kentaro Arikawa,2 Gregor Belušic,ˇ 3 and Almut Kelber4 1Faculty of Science and Engineering, University of Groningen, 9700 Groningen, The Netherlands; email: [email protected] 2Department of Evolutionary Studies of Biosystems, SOKENDAI Graduate University for Advanced Studies, Kanagawa 240-0193, Japan 3Department of Biology, Biotechnical Faculty, University of Ljubljana, 1000 Ljubljana, Slovenia; email: [email protected] 4Lund Vision Group, Department of Biology, University of Lund, 22362 Lund, Sweden; email: [email protected] Annu. Rev. Entomol. 2021. 66:23.1–23.28 Keywords The Annual Review of Entomology is online at photoreceptor, compound eye, pigment, visual pigment, behavior, opsin, ento.annualreviews.org anatomy https://doi.org/10.1146/annurev-ento-061720- 071644 Abstract Annu. Rev. Entomol. 2021.66. Downloaded from www.annualreviews.org Copyright © 2021 by Annual Reviews. Color vision is widespread among insects but varies among species, depend- All rights reserved ing on the spectral sensitivities and interplay of the participating photore- Access provided by University of New South Wales on 09/26/20. For personal use only. ceptors. The spectral sensitivity of a photoreceptor is principally determined by the absorption spectrum of the expressed visual pigment, but it can be modified by various optical and electrophysiological factors. For example, screening and filtering pigments, rhabdom waveguide properties, retinal structure, and neural processing all influence the perceived color signal. -

Region of Nymphalidae and Libytheidae (Lepidoptera) from Japan
MORPHOLOGICAL STUDIES ON THE OCCIPITAL REGION OF NYMPHALIDAE AND LIBYTHEIDAE (LEPIDOPTERA) FROM JAPAN Takashi TSUBUKI and Nagao KOYAMA * .h2monji junior High School, 10-33, Kita6tsz{ha 1, Toshima-feu, Toleyo (170), 1opan ** Biological Laboratory, Shinshi2 Udeiversity, Cllada (386), 14pan CONTENTS I. Introduction・--・ny・・・・・・・・・・・・・・・・・-・-・・・・・・-・---・--t・-・---・ny・・・・-・-・・・-・-・・-・・・1 II. Materials and methods・-・・・・・・・・・t・・・・・・t・・・・・・・・・・・・・・`・・・・・・・・・・-・・・-・・・・・・-d・・・・・・・・・・・・2 III. General morphology of the occipital region ・・・・・・}・・・・・・・・・・・・・・・・・・・・・・・・・・・・・4 IV. Occipital structure of each species in Nymphaliclae and Libytheidae ・-ib-・・・・-・・・-・ny・・・・・--・-・・・・・・・-・・-・----・b-・・・----・-・・・-・・-・--・・-6 V. Phylogenical grouping of Nymphalidae based on the occipital structure・t・・・・・・・--・--・・-・-・・・・・・・・・・-・・・・-ny-・・・ny-・--・-----・H"""""".","".."."lg VI. Summary ・・・・-・-・-・・・・・・-・・----・・・-・・・・-・-・-・-----・・-------・-・----・23 VII. Literatures cited ・・・--・・・・・・・・・`・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・--・-・・-・・・・・・tt・・・・・・23 I. INTRODUCTION A considerable number of studies have been carried out on the occipital morphology of moths since YAGI and KoYAMA's report of 1963 (KoYAMA and MlyATA, 1969, 1970, 1975;MIYATA, 1971, 1972, 1973, 1974, 1975;MIyATA and KOYAMA, 1971, 1972, 1976). In the butterfiies, however, few papers were available concerning the occipital region, on which EHRucH (1958, a b), YAGI and KoyAMA (1963) and KoyAMA and OGAwA (1972) briefly described. Then, tried to study the occipital region of butterflies, the authors observed it preliminarily (TsuBuKI et aL 1975). The present paper deals with the occipital structure of Nymphalidae and Libytheidae from Japan with reference to its bearing on systematics. Before going further, the authors wish to express their gratitude to Prof. Dr. H. SAwADA and Prof. Dr. N. GOKAN, Tokyo University of Agriculture, for their advice and assistance. Thanks are also due to Mr. N. K6DA who gave valuable materials for this work, and to Mr. Y. YAGucm and the members of JEOL Ltd. -
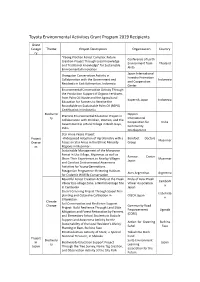
List of Previous Grant Projects
Toyota Environmental Activities Grant Program 2019 Recipients Grant Catego Theme Project Description Organization Country ry "Kaeng Krachan Forest Complex: Future Conference of Earth Creation Project Through Local Knowledge Environment from Thailand and Traditional Knowledge" for Sustainable Akita Environmental Innovation Japan International Orangutan Conservation Activity in Forestry Promotion Collaboration with the Government and Indonesia and Cooperation Residents in East Kalimantan, Indonesia Center Environmental Conservation Activity Through the Production Support of Organic Fertilizers from Palm Oil Waste and the Agricultural Kopernik Japan Indonesia Education for Farmers to Receive the Roundtable on Sustainable Palm Oil (RSPO) Certification in Indonesia Biodiversi Nippon Practical Environmental Education Project in ty International Collaboration with Children, Women, and the Cooperation for India Government in a Rural Village in Bodh Gaya, Community India Development Star Anise Peace Project Project -Widespread Adoption of Agroforestry with a Barefoot Doctors Myanmar Overse Focus on Star Anise in the Ethnic Minority Group as Regions in Myanmar- Sustainable Management of the Mangrove Forest in Uto Village, Myanmar, as well as Ramsar Center Share Their Experiences to Nearby Villages Myanmar Japan and Conduct Environmental Awareness Activities for Young Generations Patagonian Programme: Restoring Habitats Aves Argentinas Argentina for Endemic Wildlife Conservation Beautiful Forest Creation Activity at the Preah Pride of Asia: Preah -

Practical Experiences in Invasive Alien Plant Control
ROSALIA Handbooks ROSALIA Handbooks Practical Experiences in Invasive Alien Plant Control Second, revised and expanded edition Invasive plant species pose major agricultural, silvicultural, human health and ecological problems worldwide, and are considered the most signifi cant threat for nature conservation. Species invading natural areas in Hungary have been described by a number of books published in the Practical Experiences in Invasive Alien Plant Control last few years. A great amount of experience has been gathered about the control of these species in some areas, which we can read about in an increasing number of articles; however, no book has been published with regards to the whole country. Invasions affecting larger areas require high energy and cost input, and the effectiveness and successfulness of control can be infl uenced by a number of factors. The development of effective, widely applicable control and eradication technologies is preceded by experiments and examinations which are based on a lot of practical experience and often loaded with negative experiences. National park directorates, forest and agricultural managers and NGOs in many parts of Hungary are combatting the spread of invasive species; however, the exchange of information and conclusion of experiences among the managing bodies is indispensable. The aim of the present volume is to facilitate this by summarizing experiences and the methods applied in practice; which, we hope, will enable us to successfully stop the further spread of invasive plant species and effectively protect our natural values. Magyarország-Szlovákia Partnerséget építünk Határon Átnyúló Együttműködési Program 2007-2013 Duna-Ipoly National Park Directorate rrosaliaosalia kkezikonyvezikonyv 3 aangng jjav.inddav.indd 1 22017.12.15.017.12.15. -

Catalogue of the Type Specimens of Lepidoptera Rhopalocera in the Hill Museum
Original from and digitized by National University of Singapore Libraries Original from and digitized by National University of Singapore Libraries Original from and digitized by National University of Singapore Libraries Original from and digitized by National University of Singapore Libraries CATALOGUE OF THE Type Specimens of Lepidoptera Rhopalocera IN THE HILL MUSEUM BY A. G. GABRIEL, F.E.S. Issued June, 1932 LONDON JOHN BALE, SONS & DANIELSSON, LTD. 83-91, GBEAT TITCHFIELD STEEET, OXEOED STEEET, W. 1 1932 Price 20/- Original from and digitized by National University of Singapore Libraries Unfortunately Mr. Joicey did not live to see the publication of this Catalogue. It will however remain, together with the four completed volumes of the " Bulletin of the Hill Museum," as a lasting memorial to to the magnificent collection of Lepidoptera amassed by Mr. Joicey, and to the work carried out at the Hill Museum under his auspices. G. Talbot. Original from and digitized by National University of Singapore Libraries CATALOGUE OF THE TYPE SPECIMENS OF LEPIDOPTERA RHOPALOCERA IN THE HILL MUSEUM. By A. G. GABRIEL, F.E.S. INTRODUCTION BY G. TALBOT. It is important to know exactly where type specimens are to be found. The British Museum set an example by publishing catalogues of some of their Rhopalocera types, and we hope this will be continued. Mr. Gabriel, who was responsible for that work, has been asked by Mr. Joicey to prepare a catalogue for the Hill Museum. The original description of almost every name in this catalogue has been examined for the correct reference, and where the sex or habitat was wrongly quoted, the necessary correction has been made. -

Photonic Crystal Structure of Wing Scales in Sasakia Charonda Butterflies
Materials Transactions, Vol. 51, No. 2 (2010) pp. 202 to 208 Special Issue on Development and Fabrication of Advanced Materials Assisted by Nanotechnology and Microanalysis #2010 The Japan Institute of Metals Photonic Crystal Structure of Wing Scales in Sasakia Charonda Butterflies Jirˇina Mateˇjkova´-Plsˇkova´1, Dalibor Jancˇik1, Miroslav Masˇla´nˇ1, Satoshi Shiojiri2 and Makoto Shiojiri3;* 1Centre for Nanomaterial Research, Faculty of Science, Palacky University in Olomouc, Slechtitelu 11, 783 71 Olomouc, Czech Republic 2Matsubara Junior High-school, Kyoto 604-8812, Japan 3Professor Emeritus of Kyoto Institute of Technology, 1-297 Wakiyama, Kyoto 618-0091, Japan The hindwings of the male Sasakia charonda charonda butterflies comprise iridescent purple-blue areas, iridescent white-pearl areas, yellow spots and red spots as well as brown background. We have examined the microstructure of their scales by scanning electron microscopy, for applying their photonic crystal structures to fine light manipulators such as reflection elements in laser diodes. The scales in the yellow spots, red spots and brown background have almost the same structure, which is an optical diffraction grating made of ridges with two cuticle layers. Their difference comes from the contained pigments. The scales in the iridescent purple-blue and white-pearl are also the same in structure. They have seven tilted cuticle layers lapped on the ridges, which also constitute a grating. The widths of the ridge and groove in the grating are different between scales of the two kinds. It is shown that the vivid iridescence is mainly attributed to multiple interferences caused between rays reflected from the seven cuticle layers with air gaps.