Taste Receptor Tas2r5 and Tas1r3 Is Expressed in Sublingual Gland
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Modulation of Food Intake by Differential TAS2R Stimulation In
nutrients Article Modulation of Food Intake by Differential TAS2R Stimulation in Rat Carme Grau-Bové 1, Alba Miguéns-Gómez 1 , Carlos González-Quilen 1 , José-Antonio Fernández-López 2,3 , Xavier Remesar 2,3 , Cristina Torres-Fuentes 4 , Javier Ávila-Román 4 , Esther Rodríguez-Gallego 1, Raúl Beltrán-Debón 1 , M Teresa Blay 1 , Ximena Terra 1 , Anna Ardévol 1,* and Montserrat Pinent 1 1 MoBioFood Research Group, Department of Biochemistry and Biotechnology, Universitat Rovira i Virgili, 43007 Tarragona, Spain; [email protected] (C.G.-B.); [email protected] (A.M.-G.); [email protected] (C.G.-Q.); [email protected] (E.R.-G.); [email protected] (R.B.-D.); [email protected] (M.T.B.); [email protected] (X.T.); [email protected] (M.P.) 2 Department of Biochemistry and Molecular Biomedicine, Faculty of Biology, University of Barcelona, Av. Diagonal 643, 08028 Barcelona, Spain; [email protected] (J.-A.F.-L.); [email protected] (X.R.) 3 CIBER Obesity and Nutrition, Institute of Health Carlos III, Av. Diagonal 643, 08028 Barcelona, Spain 4 Nutrigenomics Research Group, Department of Biochemistry and Biotechnology, Universitat Rovira i Virgili, 43007 Tarragona, Spain; [email protected] (C.T.-F.); [email protected] (J.Á.-R.) * Correspondence: [email protected]; Tel.: +34-977-559-566 Received: 31 October 2020; Accepted: 4 December 2020; Published: 10 December 2020 Abstract: Metabolic surgery modulates the enterohormone profile, which leads, among other effects, to changes in food intake. Bitter taste receptors (TAS2Rs) have been identified in the gastrointestinal tract and specific stimulation of these has been linked to the control of ghrelin secretion. -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

G Protein‐Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: G protein-coupled receptors. British Journal of Pharmacology (2019) 176, S21–S141 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: G protein-coupled receptors Stephen PH Alexander1 , Arthur Christopoulos2 , Anthony P Davenport3 , Eamonn Kelly4, Alistair Mathie5 , John A Peters6 , Emma L Veale5 ,JaneFArmstrong7 , Elena Faccenda7 ,SimonDHarding7 ,AdamJPawson7 , Joanna L Sharman7 , Christopher Southan7 , Jamie A Davies7 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Monash Institute of Pharmaceutical Sciences and Department of Pharmacology, Monash University, Parkville, Victoria 3052, Australia 3Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK 4School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 5Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 6Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 7Centre for Discovery Brain Sciences, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. Although the Concise Guide represents approximately 400 pages, the material presented is substantially reduced compared to information and links presented on the website. -

Bivariate Genome-Wide Association Analysis Strengthens the Role of Bitter Receptor Clusters on Chromosomes 7 and 12 in Human Bitter Taste
bioRxiv preprint doi: https://doi.org/10.1101/296269; this version posted April 6, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Bivariate genome-wide association analysis strengthens the role of bitter receptor clusters on chromosomes 7 and 12 in human bitter taste Liang-Dar Hwang1,2,3,4, Puya Gharahkhani1, Paul A. S. Breslin5,6, Scott D. Gordon1, Gu Zhu1, Nicholas G. Martin1, Danielle R. Reed5, and Margaret J. Wright2,7 1 QIMR Berghofer Medical Research Institute, Herston, Queensland 4006, Australia 2 Queensland Brain Institute, University of Queensland, St Lucia, Queensland 4072, Australia 3 Faculty of Medicine, University of Queensland, Herston, Queensland 4006, Australia 4 University of Queensland Diamantina Institute, University of Queensland, Translational Research Institute, Woolloongabba, Queensland 4102, Australia 5 Monell Chemical Senses Center, Philadelphia, Pennsylvania 19104, USA 6 Department of Nutritional Sciences, School of Environmental and Biological Sciences, Rutgers University, New Brunswick NJ, 08901 USA 7 Centre for Advanced Imaging, University of Queensland, St Lucia, Queensland 4072, Australia Correspondence to be sent to: Liang-Dar Hwang University of Queensland Diamantina Institute Wolloongabba QLD 4102, Australia Email: [email protected] Telephone: +61 7 3443 7976 Fax: +61 7 3443 6966 1 bioRxiv preprint doi: https://doi.org/10.1101/296269; this version posted April 6, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

The Bitter Taste Receptor Tas2r14 Is Expressed in Ovarian Cancer and Mediates Apoptotic Signalling
THE BITTER TASTE RECEPTOR TAS2R14 IS EXPRESSED IN OVARIAN CANCER AND MEDIATES APOPTOTIC SIGNALLING by Louis T. P. Martin Submitted in partial fulfilment of the requirements for the degree of Master of Science at Dalhousie University Halifax, Nova Scotia June 2017 © Copyright by Louis T. P. Martin, 2017 DEDICATION PAGE To my grandparents, Christina, Frank, Brenda and Bernie, and my parents, Angela and Tom – for teaching me the value of hard work. ii TABLE OF CONTENTS LIST OF TABLES ............................................................................................................. vi LIST OF FIGURES .......................................................................................................... vii ABSTRACT ....................................................................................................................... ix LIST OF ABBREVIATIONS AND SYMBOLS USED .................................................... x ACKNOWLEDGEMENTS .............................................................................................. xii CHAPTER 1 INTRODUCTION ........................................................................................ 1 1.1 G-PROTEIN COUPLED RECEPTORS ................................................................ 1 1.2 GPCR CLASSES .................................................................................................... 4 1.3 GPCR SIGNALING THROUGH G PROTEINS ................................................... 6 1.4 BITTER TASTE RECEPTORS (TAS2RS) ........................................................... -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 7, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on September 29, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009. -

Clinical, Molecular, and Immune Analysis of Dabrafenib-Trametinib
Supplementary Online Content Chen G, McQuade JL, Panka DJ, et al. Clinical, molecular and immune analysis of dabrafenib-trametinib combination treatment for metastatic melanoma that progressed during BRAF inhibitor monotherapy: a phase 2 clinical trial. JAMA Oncology. Published online April 28, 2016. doi:10.1001/jamaoncol.2016.0509. eMethods. eReferences. eTable 1. Clinical efficacy eTable 2. Adverse events eTable 3. Correlation of baseline patient characteristics with treatment outcomes eTable 4. Patient responses and baseline IHC results eFigure 1. Kaplan-Meier analysis of overall survival eFigure 2. Correlation between IHC and RNAseq results eFigure 3. pPRAS40 expression and PFS eFigure 4. Baseline and treatment-induced changes in immune infiltrates eFigure 5. PD-L1 expression eTable 5. Nonsynonymous mutations detected by WES in baseline tumors This supplementary material has been provided by the authors to give readers additional information about their work. © 2016 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/30/2021 eMethods Whole exome sequencing Whole exome capture libraries for both tumor and normal samples were constructed using 100ng genomic DNA input and following the protocol as described by Fisher et al.,3 with the following adapter modification: Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter. In-solution hybrid selection was performed using the Illumina Rapid Capture Exome enrichment kit with 38Mb target territory (29Mb baited). The targeted region includes 98.3% of the intervals in the Refseq exome database. Dual-indexed libraries were pooled into groups of up to 96 samples prior to hybridization. -

Role of Bitter Taste Receptors in Regulating Gastric Accommodation in Guinea Pigs
1521-0103/369/3/466–472$35.00 https://doi.org/10.1124/jpet.118.256008 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 369:466–472, June 2019 Copyright ª 2019 The Author(s). This is an open access article distributed under the CC BY Attribution 4.0 International license. Role of Bitter Taste Receptors in Regulating Gastric Accommodation in Guinea Pigs Yumi Harada, Junichi Koseki, Hitomi Sekine, Naoki Fujitsuka, and Hiroyuki Kobayashi Tsumura Kampo Research Laboratories, Tsumura & Co., Ibaraki, Japan (Y.H., J.K., H.S., N.F.) and Center for Advanced Kampo Medicine and Clinical Research, Juntendo Graduate School of Medicine, Tokyo, Japan (H.K.) Received January 10, 2019; accepted April 4, 2019 ABSTRACT Taste stimulants play important roles in triggering digestion and the consequent intrabag pressure in the proximal stomach Downloaded from absorption of nutrients and in toxin detection, under the control of guinea pigs. Effects of the Kampo medicine rikkunshito of the gut-brain axis. Bitter compounds regulate gut hormone (RKT) and its bitter components liquiritigenin and naringenin secretion and gastrointestinal motility through bitter taste on GA were also examined. Intraoral DB (0.2 nmol/ml) admin- receptors (TAS2Rs) located in the taste buds on the tongue istration enhanced GA. Intragastric DB administration (0.1 and and in the enteroendocrine cells. Gastric accommodation 1 nmol/kg) promoted GA, whereas higher DB doses (30 mmol/kg) (GA) is an important physiologic function. However, the role inhibited it. Similar changes in GA were observed with intra- of TAS2R agonists in regulating GA remains unclear. -
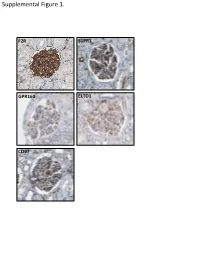
Supplemental Data
Supplemental Figure 1. F2R S1PR1 GPR160 ELTD1 CD97 Supplemental Figure 2. A B brain heart lung liver kidney spleen testis skm Expression in mouse ssues glom rok Gprc5a Gapdh Human Protein Atlas RNAseq database Supplemental Figure 3. A Gprc5a Pdgfrb Merged CL CL Gprc5a CD31 Merged CL CL B C 1.4 * 1.2 mc 1 matrix 0.8 0.6 0.4 matrix 0.2 0 pod end 1 2 mes3 D Vector Gprc5a E 40 kD - - Gprc5a - acn Supplemental Figure 4. Control 12 month-old KO 12 month-old A B C D E F 1 2 0.8 * 1.5 score 0.6 m µ 1 0.4 0.2 Slits/ 0.5 Mesangial 0 0 Ctrl KO Ctrl KO Supplemental Figure 5. A B 1.2 Vector Gprc5a 1 * 0.8 0.6 * 0.4 * Normalized Density 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 C 1.8 siCON 1.6 siGprc5a * 1.4 * * 1.2 1 0.8 0.6 NormalizedDensity 0.4 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 Supplemental table 1. List of glomerulus-expressed GPCRs as detected by qPCR. Data shown as mean ± standard deviation (Glom=glomerulus, Rok=rest of kidney). GPCR Glom Rok Glom/Rok LPAR6 41892,11 ± 38478,89 1040,06 ± 1370,12 39,28 ELTD1 30275,64 ± 14085,26 33,23 ± 46,99 910,13 GPR116 24020,06 ± 7789,84 55,1 ± 47,75 434,96 PTH1R 15402,81 ± 17644,32 7521,01 ± 3264,57 1,05 CALCRL 14096,09 ± 3854,84 199,06 ± 222,52 69,81 HPRT1 13342,04 ± 10677,69 1824,77 ± 1767,23 6,31 S1PR5 9474,7 ± 10124,3 110,84 ± 29,54 84,48 LPHN2 8645,89 ± 914,74 256,04 ± 293,87 32,77 FZD1 8176,2 ± 4947,45 1321,97 ± 1311,91 5,18 CXCR4 7097,31 ± 4388,91 535,98 ± 640,28 12,24 GPR160 6446,59 ± 1550,07 4816,3 ± 5918,99 0,34 NPY1R 6177,74 ± 6282,76 209,64 ± 296,48 28,47 PTGER4 5323,66 ± 3789,51 179,13 ± 210 28,72 RXFP1 -

G Protein‐Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: G protein-coupled receptors. British Journal of Pharmacology (2019) 176, S21–S141 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: G protein-coupled receptors Stephen PH Alexander1 , Arthur Christopoulos2 , Anthony P Davenport3 , Eamonn Kelly4, Alistair Mathie5 , John A Peters6 , Emma L Veale5 ,JaneFArmstrong7 , Elena Faccenda7 ,SimonDHarding7 ,AdamJPawson7 , Joanna L Sharman7 , Christopher Southan7 , Jamie A Davies7 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Monash Institute of Pharmaceutical Sciences and Department of Pharmacology, Monash University, Parkville, Victoria 3052, Australia 3Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK 4School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 5Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 6Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 7Centre for Discovery Brain Sciences, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. Although the Concise Guide represents approximately 400 pages, the material presented is substantially reduced compared to information and links presented on the website.