A Bitter Taste in Your Heart
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Database Integrating Genomic Indel Polymorphisms That Distinguish Mouse Strains Keiko Akagi1,2, Robert M
D600–D606 Nucleic Acids Research, 2010, Vol. 38, Database issue Published online 20 November 2009 doi:10.1093/nar/gkp1046 MouseIndelDB: a database integrating genomic indel polymorphisms that distinguish mouse strains Keiko Akagi1,2, Robert M. Stephens3, Jingfeng Li2,4, Evgenji Evdokimov5, Michael R. Kuehn5, Natalia Volfovsky3 and David E. Symer2,4,6,7,8,9,* 1Mouse Cancer Genetics Program, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, 2Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University Comprehensive Cancer Center, Columbus, OH 43210, 3Advanced Biomedical Computing Center, Information Systems Program, SAIC-Frederick, Inc., NCI-Frederick, Frederick, MD 21702, 4Basic Research Laboratory, 5Laboratory of Protein Dynamics and Signaling, 6Laboratory of Biochemistry and Molecular Biology, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, 7Human Cancer Genetics Program, 8Departments of Internal Medicine and 9Biomedical Informatics, The Ohio State University Comprehensive Cancer Center, Columbus, OH 43210, USA Received August 5, 2009; Revised October 23, 2009; Accepted October 27, 2009 ABSTRACT wide-ranging functional differences. This extensive phenotypic variation has helped to make the mouse a MouseIndelDB is an integrated database resource premier model organism, mimicking many aspects of containing thousands of previously unreported human diversity and diseases. Understanding the mouse genomic indel (insertion and deletion) genomic differences that -

Pathway in Protective Immunity Jeremy Manry, Guillaume Laval, Etienne Patin, Simona Fornarino, Christiane Bouchier, Magali Tichit, Luis Barreiro, Lluis Quintana-Murci
Evolutionary genetics evidence of an essential, non-redundant role of the IFN-γ pathway in protective immunity Jeremy Manry, Guillaume Laval, Etienne Patin, Simona Fornarino, Christiane Bouchier, Magali Tichit, Luis Barreiro, Lluis Quintana-Murci To cite this version: Jeremy Manry, Guillaume Laval, Etienne Patin, Simona Fornarino, Christiane Bouchier, et al.. Evo- lutionary genetics evidence of an essential, non-redundant role of the IFN-γ pathway in protective immunity. Human Mutation, Wiley, 2011, 32 (6), pp.633-42. 10.1002/humu.21484. hal-00627541 HAL Id: hal-00627541 https://hal.archives-ouvertes.fr/hal-00627541 Submitted on 29 Sep 2011 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Human Mutation Evolutionary genetics evidence of an essential, non- redundant role of the IFN-γ pathway in protective immunity For Peer Review Journal: Human Mutation Manuscript ID: humu-2010-0462.R1 Wiley - Manuscript type: Research Article Date Submitted by the 23-Jan-2011 Author: Complete List of Authors: Manry, Jeremy; Institut Pasteur Laval, Guillaume; Institut Pasteur Patin, Etienne; Institut Pasteur Fornarino, Simona; Institut Pasteur Bouchier, Christiane; Institut Pasteur Tichit, Magali; Institut Pasteur Barreiro, Luis; University of Chicago Quintana-Murci, Lluis; Institut Pasteur, EEMI IFNG, IFNGR1, IFNGR2, natural selection, polymorphism, Key Words: population genetics John Wiley & Sons, Inc. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcriptomic Analysis of Native Versus Cultured Human and Mouse Dorsal Root Ganglia Focused on Pharmacological Targets Short
bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Transcriptomic analysis of native versus cultured human and mouse dorsal root ganglia focused on pharmacological targets Short title: Comparative transcriptomics of acutely dissected versus cultured DRGs Andi Wangzhou1, Lisa A. McIlvried2, Candler Paige1, Paulino Barragan-Iglesias1, Carolyn A. Guzman1, Gregory Dussor1, Pradipta R. Ray1,#, Robert W. Gereau IV2, # and Theodore J. Price1, # 1The University of Texas at Dallas, School of Behavioral and Brain Sciences and Center for Advanced Pain Studies, 800 W Campbell Rd. Richardson, TX, 75080, USA 2Washington University Pain Center and Department of Anesthesiology, Washington University School of Medicine # corresponding authors [email protected], [email protected] and [email protected] Funding: NIH grants T32DA007261 (LM); NS065926 and NS102161 (TJP); NS106953 and NS042595 (RWG). The authors declare no conflicts of interest Author Contributions Conceived of the Project: PRR, RWG IV and TJP Performed Experiments: AW, LAM, CP, PB-I Supervised Experiments: GD, RWG IV, TJP Analyzed Data: AW, LAM, CP, CAG, PRR Supervised Bioinformatics Analysis: PRR Drew Figures: AW, PRR Wrote and Edited Manuscript: AW, LAM, CP, GD, PRR, RWG IV, TJP All authors approved the final version of the manuscript. 1 bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -
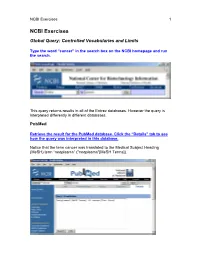
NCBI Exercises 1
NCBI Exercises 1 NCBI Exercises Global Query: Controlled Vocabularies and Limits Type the word “cancer” in the search box on the NCBI homepage and run the search. This query returns results in all of the Entrez databases. However the query is interpreted differently in different databases. PubMed Retrieve the result for the PubMed database. Click the “Details” tab to see how the query was interpreted in this database. Notice that the term cancer was translated to the Medical Subject Heading (MeSH) term “neoplasms” ("neoplasms"[MeSH Terms]). NCBI Exercises 2 MeSH is a controlled vocabulary that is used to index all articles in PubMed. In the details box, edit the query to remove the portion that searched for cancer as a text word and run the search. Notice that the number of articles retrieved has changed. These will be a more relevant set of results. You can force the PubMed engine to only search the MeSH vocabulary or specify any other indexed field through the “Limits” tab. Use the Web browser’s back button to return to the Global query page and retrieve the PubMed results again. Now click on the “Limits” tab. Select “MeSH terms” from the first drop-down menu, the one headed by “All Fields”. Now run the search with the limit in place and check the “Details” tab to verify that only the MeSH term translation was used. Nucleotide Use the Web browser’s back button to return to the Global query page. Retrieve the results for the nucleotide database. Click the “Details” tab to see how the query was interpreted for this molecular database. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Taste Receptor Tas2r5 and Tas1r3 Is Expressed in Sublingual Gland
inal chem ic is d t Li et al., Med Chem (Los Angeles) 2018, 8:5 e r M y Medicinal Chemistry DOI: 10.4172/2161-0444.1000504 ISSN: 2161-0444 t Research Article Open Access Taste Receptor Tas2r5 and Tas1r3 is Expressed in Sublingual Gland Feng Li*, Bowen Niu and Mengmin Zhu Shanghai Public Health Clinical Center, Fudan University, Shanghai 201508, China Abstract Type 2 taste receptors (TAS2Rs), a large family of GPCRs, were first discovered in the gustatory system, and are co-expressed in a subset of taste receptor cells and detect bitter-tasting compounds. Type 1 taste receptors (TAS1R3) function as an obligate partner for both the umami receptor and the sweet receptor. Recently, it has become clear that taste receptors are also expressed outside the gustatory system. Here, with Tas2r5-Cre/GFP and Tas1r3-Cre/GFP transgenic mice, the expression of taste receptors (Tas1r3 or Tas2r5) is observed in serous gland and mucous gland of tongue. Taste signal transduction cascade (Gnat3 and PLC-β2) is also detected in serous gland and mucous gland of tongue. After DTA expression in Tas2r5+ and Tas1r3+ cells, the expression of taste receptors and taste signal transduction cascade is ablated in serous and mucous gland of tongue. Keywords: Serous gland; Mucous gland; Taste receptor; Tongue; their associated G-protein genes has also been reported in mammalian Transgenic mice brain, indicating that the Tas1r2/Tas1r3 is a candidate membrane- bound brain glucosensor [18,19]. Introduction In previous study, we generate two transgenic mice with the It is believed that taste is the sensory modality that guides organisms modified BAC-DNA, in which a fusion protein of GFP/Cre is driven to recognize and consume digestible material and avoid toxins. -

Molecular Evolution and Deorphanization of Bitter Taste Receptors in a Vampire Bat
Integrative Zoology 2020; 0: 1–11 doi: 10.1111/1749-4877.12509 ORIGINAL ARTICLE Molecular evolution and deorphanization of bitter taste receptors in a vampire bat Qin LU,1 Hengwu JIAO,1 Yi WANG,1 Ngawang NORBU2 and Huabin ZHAO1,2 1Department of Ecology, Tibetan Centre for Ecology and Conservation at WHU-TU, Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan, China and 2Research Center for Ecology, College of Science, Tibet University, Lhasa, China Abstract Bats represent the largest dietary radiation in a single mammalian order, and have become an emerging model group for studying dietary evolution. Taste receptor genes have proven to be molecular signatures of dietary diversification in bats. For example, all 3 extant species of vampire bats have lost many bitter taste receptor genes (Tas2rs) in association with their dietary shift from insectivory to sanguivory. Indeed, only 8 full-length Tas2rs were identified from the high-quality genome of the common vampire bat (Desmodus rotundus). However, it is presently unknown whether these bitter receptors are functional, since the sense of taste is less important in vampire bats, which have an extremely narrow diet and rely on other senses for acquiring food. Here, we applied a molecular evolutionary analysis of Tas2rs in the common vampire bat compared with non-vampire bats. Furthermore, we provided the first attempt to deorphanize all bitter receptors of the vampire bat using a cell-based assay. We found that all Tas2r genes in the vampire bat have a level of selective pressure similar to that in non-vampire bats, suggesting that this species must have retained some bitter taste functions. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Transcriptional Profiling Identifies the Lncrna PVT1 As a Novel Regulator of the Asthmatic Phenotype in Human Airway Smooth Muscle
Accepted Manuscript Transcriptional profiling identifies the lncRNA PVT1 as a novel regulator of the asthmatic phenotype in human airway smooth muscle Philip J. Austin, MSc, Eleni Tsitsiou, PhD, Charlotte Boardman, MD, Simon W. Jones, PhD, Mark A. Lindsay, PhD, Ian M. Adcock, PhD, Kian Fan Chung, MD PhD, Mark M. Perry, PhD PII: S0091-6749(16)30571-1 DOI: 10.1016/j.jaci.2016.06.014 Reference: YMAI 12203 To appear in: Journal of Allergy and Clinical Immunology Received Date: 5 April 2016 Revised Date: 24 May 2016 Accepted Date: 13 June 2016 Please cite this article as: Austin PJ, Tsitsiou E, Boardman C, Jones SW, Lindsay MA, Adcock IM, Chung KF, Perry MM, Transcriptional profiling identifies the lncRNA PVT1 as a novel regulator of the asthmatic phenotype in human airway smooth muscle, Journal of Allergy and Clinical Immunology (2016), doi: 10.1016/j.jaci.2016.06.014. This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT 1 Transcriptional profiling identifies the lncRNA PVT1 as a novel 2 regulator of the asthmatic phenotype in human airway smooth muscle 3 4 Philip J. Austin MSc 1, Eleni Tsitsiou PhD 2, Charlotte Boardman MD 1, Simon W. -

Supplementary Tables Supplemental Table S1: Comparison of the Coverage of Reference Panels Used for SNP Imputation. the Markers
Supplementary Tables Supplemental Table S1: Comparison of the coverage of reference panels used for SNP imputation. The markers on the Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) reference panel and Haplotype Reference Consortium (HRG) used to impute SNPs from our AA and CAU participants were compared to one thousand genomes (1kG) dataset. Loci information CAAPA vs 1kG HRC vs 1kG Total loci 45,639,158 90,558,388 Overlapping loci 24,880,301 49,826,569 Percent overlap 54.52% 55.02% 1kG-only loci 9,363,544 15,148,191 Ref-only loci 8,461,186 22,649,501 Supplemental Table S2: SNP imputation results. The total number of SNPs imputed for the AA and CAU participants either using the Michigan imputation server (Minimac) or Beagle. Targets prepared for: AA CAU Beagle 730,616 726,165 Minimac 698,343 660,733 Supplemental Table S3: Allele frequencies of TA2R38 SNPs by each ancestral group and time point. The rs number of each SNP, the location of SNP buy chromosome (CHR) and base pair position (POS) is provided along with the allele frequency (ALLELE:FREQ) for each SNP. Baseline 6-month Baseline 6-month AA (N = 297) AA (N = 234) CAU (N = 198) CAU (N = 151) SNP CHR POS ALLELE:FREQ ALLELE:FREQ ALLELE:FREQ ALLELE:FREQ rs10246939 7 141672604 T:0.49 C:0.51 T:0.50 C:0.50 T:0.54 C:0.46 T:0.54 C:0.46 rs1726866 7 141672705 G:0.68 A:0.32 G:0.68 A:0.32 G:0.46 A:0.54 G:0.46 A:0.54 rs713598 7 141673345 C:0.50 G:0.50 C:0.50 G:0.50 C:0.58 G:0.42 C:0.58 G:0.42 Supplemental Table S4: Linkage disequilibrium analysis of TAS2R38 SNPs at each time point of the intervention. -

(12) United States Patent (10) Patent No.: US 9,347,934 B2 Shekdar Et Al
USOO9347934B2 (12) United States Patent (10) Patent No.: US 9,347,934 B2 Shekdar et al. (45) Date of Patent: May 24, 2016 (54) ASSAYS FOR IDENTIFYING COMPOUNDS 2008, OO38739 A1 2/2008 Li et al. THAT MODULATE BITTERTASTE 2008/0167286 A1* 7/2008 Gopalakrishnan et al. ........................ 514,21016 (71) Applicants: CHROMOCELL CORPORATION, 2010/01298.33 A1* 5/2010 Brune et al. ................. 435/721 North Brunswick, NJ (US); KRAFT FOODS GROUP BRANDS LLC, FOREIGN PATENT DOCUMENTS Northfield, IL (US) CN 1341632 A 3, 2002 CN 101583717 A 11, 2009 (72) Inventors: Kambiz Shekdar, New York, NY (US); CN 101828.111 A 9, 2010 Purvi Manoj Shah, Bridgewater, NJ WO WO-0038536 A2 7, 2000 WO WO-2004O29087 4/2004 (US); Joseph Gunnet, Flemington, NJ WO WO-2006053771 A2 5, 2006 (US); Jane V. Leland, Wilmette, IL WO WO-2007002026 A2 1/2007 (US); Peter H. Brown, Glenview, IL WO WO-2008057470 5, 2008 (US); Louise Slade, Morris Plains, NJ WO WO-2008119.195 A1 10, 2008 (US) WO WO-20081191.96 10, 2008 WO WO-20081191.97 10, 2008 (73) Assignees: Chromocell Corporation, North W WSi. A2 1929 Brunswick, NJ (US); Kraft Foods WO WO-2010O886.33 8, 2010 Group Brands LLC, Northfield, IL WO WO-2010O99983 A1 9, 2010 (US) WO WO-2013022947 2, 2013 (*) Notice: Subject to any disclaimer, the term of this OTHER PUBLICATIONS patent is extended or adjusted under 35 U.S.C. 154(b) by 0 days. Bachmanov et al., Taste Receptor Genes, 2007, 27:389-414.* Behrens et al., Structural Requirements for Bitter Taste Receptor (21) Appl.