The Deep-Sea Teleost Cornea: a Comparative Study of Gadiform Fishes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Exon Probe Sets and Bioinformatics Pipelines for All Levels of Fish Phylogenomics
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.18.949735; this version posted February 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Exon probe sets and bioinformatics pipelines for all levels of fish phylogenomics 2 3 Lily C. Hughes1,2,3,*, Guillermo Ortí1,3, Hadeel Saad1, Chenhong Li4, William T. White5, Carole 4 C. Baldwin3, Keith A. Crandall1,2, Dahiana Arcila3,6,7, and Ricardo Betancur-R.7 5 6 1 Department of Biological Sciences, George Washington University, Washington, D.C., U.S.A. 7 2 Computational Biology Institute, Milken Institute of Public Health, George Washington 8 University, Washington, D.C., U.S.A. 9 3 Department of Vertebrate Zoology, National Museum of Natural History, Smithsonian 10 Institution, Washington, D.C., U.S.A. 11 4 College of Fisheries and Life Sciences, Shanghai Ocean University, Shanghai, China 12 5 CSIRO Australian National Fish Collection, National Research Collections of Australia, 13 Hobart, TAS, Australia 14 6 Sam Noble Oklahoma Museum of Natural History, Norman, O.K., U.S.A. 15 7 Department of Biology, University of Oklahoma, Norman, O.K., U.S.A. 16 17 *Corresponding author: Lily C. Hughes, [email protected]. 18 Current address: Department of Organismal Biology and Anatomy, University of Chicago, 19 Chicago, IL. 20 21 Keywords: Actinopterygii, Protein coding, Systematics, Phylogenetics, Evolution, Target 22 capture 23 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.18.949735; this version posted February 19, 2020. -

Transcriptome Ortholog Alignment Sequence Tools (TOAST) for Phylogenomic Dataset Assembly
Transcriptome Ortholog Alignment Sequence Tools (TOAST) for Phylogenomic Dataset Assembly Dustin J. Wcisel North Carolina State University J. Thomas Howard North Carolina State University Jeffrey A. Yoder North Carolina State University Alex Dornburg ( [email protected] ) NC Museum of Natural Sciences https://orcid.org/0000-0003-0863-2283 Software Keywords: BUSCO ortholog assembly, Cetacean and teleost sh phylogeny, Missing Data Visualization, Transcriptome, Concatenated Alignment Posted Date: March 12th, 2020 DOI: https://doi.org/10.21203/rs.2.16269/v4 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at BMC Evolutionary Biology on March 30th, 2020. See the published version at https://doi.org/10.1186/s12862-020-01603-w. Page 1/18 Abstract Background Advances in next-generation sequencing technologies have reduced the cost of whole transcriptome analyses, allowing characterization of non-model species at unprecedented levels. The rapid pace of transcriptomic sequencing has driven the public accumulation of a wealth of data for phylogenomic analyses, however lack of tools aimed towards phylogeneticists to eciently identify orthologous sequences currently hinders effective harnessing of this resource. Results We introduce TOAST, an open source R software package that can utilize the ortholog searches based on the software Benchmarking Universal Single-Copy Orthologs (BUSCO) to assemble multiple sequence alignments of orthologous loci from transcriptomes for any group of organisms. By streamlining search, query, and alignment, TOAST automates the generation of locus and concatenated alignments, and also presents a series of outputs from which users can not only explore missing data patterns across their alignments, but also reassemble alignments based on user-dened acceptable missing data levels for a given research question. -

Fisheries Overview, Including Mixed-Fisheries Considerations
ICES Fisheries Overviews Bay of Biscay and Iberian Coast ecoregion Published 30 November 2020 Version 2: 3 December 2020 6.2 Bay of Biscay and Iberian Coast ecoregion – Fisheries overview, including mixed-fisheries considerations Table of contents Executive summary .................................................................................................................................................................. 1 Definition of the ecoregion ...................................................................................................................................................... 1 Mixed-fisheries considerations Bay of Biscay .......................................................................................................................... 2 Mixed-fisheries considerations Iberian waters ...................................................................................................................... 10 Who is fishing ........................................................................................................................................................................ 18 Catches over time .................................................................................................................................................................. 21 Description of the fisheries .................................................................................................................................................... 23 Fisheries management measures ......................................................................................................................................... -

Molecular Systematics of Gadid Fishes: Implications for the Biogeographic Origins of Pacific Species
Color profile: Disabled Composite Default screen 19 Molecular systematics of gadid fishes: implications for the biogeographic origins of Pacific species Steven M. Carr, David S. Kivlichan, Pierre Pepin, and Dorothy C. Crutcher Abstract: Phylogenetic relationships among 14 species of gadid fishes were investigated with portions of two mitochondrial DNA (mtDNA) genes, a 401 base pair (bp) segment of the cytochrome b gene, and a 495 bp segment of the cytochrome oxidase I gene. The molecular data indicate that the three species of gadids endemic to the Pacific Basin represent simultaneous invasions by separate phylogenetic lineages. The Alaskan or walleye pollock (Theragra chalcogramma) is about as closely related to the Atlantic cod (Gadus morhua) as is the Pacific cod (Gadus macrocephalus), which suggests that T. chalcogramma and G. macrocephalus represent separate invasions of the Pacific Basin. The Pacific tomcod (Microgadus proximus) is more closely related to the Barents Sea navaga (Eleginus navaga) than to the congeneric Atlantic tomcod (Microgadus tomcod), which suggests that the Pacific species is derived from the Eleginus lineage and that Eleginus should be synonymized with Microgadus. Molecular divergences between each of the three endemic Pacific species and their respective closest relatives are similar and consistent with contemporaneous speciation events following the reopening of the Bering Strait ca. 3.0–3.5 million years BP. In contrast, the Greenland cod (Gadus ogac) and the Pacific cod have essentially identical mtDNA sequences; differences between them are less than those found within G. morhua. The Greenland cod appears to represent a contemporary northward and eastward range extension of the Pacific cod, and should be synonymized with it as G. -

Diverse Deep-Sea Anglerfishes Share a Genetically Reduced Luminous
RESEARCH ARTICLE Diverse deep-sea anglerfishes share a genetically reduced luminous symbiont that is acquired from the environment Lydia J Baker1*, Lindsay L Freed2, Cole G Easson2,3, Jose V Lopez2, Dante´ Fenolio4, Tracey T Sutton2, Spencer V Nyholm5, Tory A Hendry1* 1Department of Microbiology, Cornell University, New York, United States; 2Halmos College of Natural Sciences and Oceanography, Nova Southeastern University, Fort Lauderdale, United States; 3Department of Biology, Middle Tennessee State University, Murfreesboro, United States; 4Center for Conservation and Research, San Antonio Zoo, San Antonio, United States; 5Department of Molecular and Cell Biology, University of Connecticut, Storrs, United States Abstract Deep-sea anglerfishes are relatively abundant and diverse, but their luminescent bacterial symbionts remain enigmatic. The genomes of two symbiont species have qualities common to vertically transmitted, host-dependent bacteria. However, a number of traits suggest that these symbionts may be environmentally acquired. To determine how anglerfish symbionts are transmitted, we analyzed bacteria-host codivergence across six diverse anglerfish genera. Most of the anglerfish species surveyed shared a common species of symbiont. Only one other symbiont species was found, which had a specific relationship with one anglerfish species, Cryptopsaras couesii. Host and symbiont phylogenies lacked congruence, and there was no statistical support for codivergence broadly. We also recovered symbiont-specific gene sequences from water collected near hosts, suggesting environmental persistence of symbionts. Based on these results we conclude that diverse anglerfishes share symbionts that are acquired from the environment, and *For correspondence: that these bacteria have undergone extreme genome reduction although they are not vertically [email protected] (LJB); transmitted. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Anglerfish Ageing Guide
ANGLERFISH AGEING GUIDE by Duarte, R., Landa, J., Quincoces, I., Dupouy, H., Bilbao, E., Dimeet, J., Marçal, A., McCormick, H. and Ni Chonchuir, G. Summary The objective of the present ageing guide is to make a compilation of all the information necessary to age the two species of European anglerfish (Lophius piscatorius and L. budegassa). The used calcified structure is the first dorsal fin ray (illicium). Anglerfish ageing is generally recognised as a difficult task. Hervé Dupouy, from IFREMER, Lorient (France), started to implement in the eighties a routine ageing procedure, based on illicia transversal sections. After recognising the benefits of this procedure and the clearer annual ring identification, compared to otoliths, researchers from Spain and Portugal followed Hervé’s work, in order to provide annual data for stock assessment. Since the beginning of the nineties, four illicia ageing workshops were held in order to improve methodologies and uniformity in the ageing criteria. This ageing guide results from the work developed during these workshops and the objective is to present all the necessary information to age anglerfish. This way, the introduction in section 1 makes a summary of the main biological particularities of these species, section 2 contains all the methodology to obtain illicia transversal sections, section 3 describes the ageing criteria and in section 4 are illicia images with marked annual rings. Participants (By alphabetical order of last name) Eli Bilbao AZTI Spain Joel Dimeet IFREMER France Rafael Duarte (coordinator) IPIMAR Portugal Hervé Dupouy IFREMER France Jorge Landa (coordinator) IEO Spain António Marçal IPIMAR Portugal Helen McCormick Marine Institute Ireland Grainne Ni Chonchuir Marine Institute Ireland Iñaki Quincoces (coordinator) AZTI Spain 2 Contents 1. -
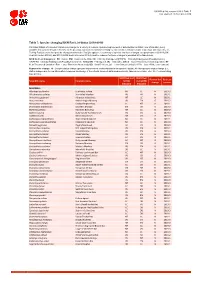
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Humboldt Bay Fishes
Humboldt Bay Fishes ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> ·´¯`·._.·´¯`·.. ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> Acknowledgements The Humboldt Bay Harbor District would like to offer our sincere thanks and appreciation to the authors and photographers who have allowed us to use their work in this report. Photography and Illustrations We would like to thank the photographers and illustrators who have so graciously donated the use of their images for this publication. Andrey Dolgor Dan Gotshall Polar Research Institute of Marine Sea Challengers, Inc. Fisheries And Oceanography [email protected] [email protected] Michael Lanboeuf Milton Love [email protected] Marine Science Institute [email protected] Stephen Metherell Jacques Moreau [email protected] [email protected] Bernd Ueberschaer Clinton Bauder [email protected] [email protected] Fish descriptions contained in this report are from: Froese, R. and Pauly, D. Editors. 2003 FishBase. Worldwide Web electronic publication. http://www.fishbase.org/ 13 August 2003 Photographer Fish Photographer Bauder, Clinton wolf-eel Gotshall, Daniel W scalyhead sculpin Bauder, Clinton blackeye goby Gotshall, Daniel W speckled sanddab Bauder, Clinton spotted cusk-eel Gotshall, Daniel W. bocaccio Bauder, Clinton tube-snout Gotshall, Daniel W. brown rockfish Gotshall, Daniel W. yellowtail rockfish Flescher, Don american shad Gotshall, Daniel W. dover sole Flescher, Don stripped bass Gotshall, Daniel W. pacific sanddab Gotshall, Daniel W. kelp greenling Garcia-Franco, Mauricio louvar -

For Creative Minds
For Creative Minds The For Creative Minds educational section may be photocopied or printed from our website by the owner of this book for educational, non-commercial uses. Cross-curricular teaching activities, interactive quizzes, and more are available online. Go to ArbordalePublishing.com and click on the book’s cover to explore all the links. Deep Ocean Habitats Things change the deeper you go in the ocean: light disappears, temperatures grow increasingly colder, and pressure gets much higher. The amount of oxygen in the water sunlight zone decreases with depth but then gets higher again at the bottom! Because these changes twilight zone affect the types of organisms that can survive there, the ocean is divided into five layers by depth called life zones. Only the sunlight zone receives enough sunlight for algae to convert light into energy midnight zone (photosynthesis). Because almost all food webs start with plants or algae, this is the zone where the most animals live. The twilight zone still gets some sunlight, but not enough for photosynthesis. The animals that live here either travel to the sunlight zone to feed or depend on food falling from above. There is no light in the midnight zone. Most abyssal zone of the animals that live here produce their own light through bioluminescence. The abyssal zone is pitch black, almost freezing cold, and has little oxygen and incredibly high pressure, yet animals still live here. In the deep trenches is the hadal zone. It is like the abyssal zone, except with even more hadal zone immense -

A Checklist of the Fishes of the Monterey Bay Area Including Elkhorn Slough, the San Lorenzo, Pajaro and Salinas Rivers
f3/oC-4'( Contributions from the Moss Landing Marine Laboratories No. 26 Technical Publication 72-2 CASUC-MLML-TP-72-02 A CHECKLIST OF THE FISHES OF THE MONTEREY BAY AREA INCLUDING ELKHORN SLOUGH, THE SAN LORENZO, PAJARO AND SALINAS RIVERS by Gary E. Kukowski Sea Grant Research Assistant June 1972 LIBRARY Moss L8ndillg ,\:Jrine Laboratories r. O. Box 223 Moss Landing, Calif. 95039 This study was supported by National Sea Grant Program National Oceanic and Atmospheric Administration United States Department of Commerce - Grant No. 2-35137 to Moss Landing Marine Laboratories of the California State University at Fresno, Hayward, Sacramento, San Francisco, and San Jose Dr. Robert E. Arnal, Coordinator , ·./ "':., - 'I." ~:. 1"-"'00 ~~ ~~ IAbm>~toriesi Technical Publication 72-2: A GI-lliGKL.TST OF THE FISHES OF TtlE MONTEREY my Jl.REA INCLUDING mmORH SLOUGH, THE SAN LCRENZO, PAY-ARO AND SALINAS RIVERS .. 1&let~: Page 14 - A1estria§.·~iligtro1ophua - Stone cockscomb - r-m Page 17 - J:,iparis'W10pus." Ribbon' snailt'ish - HE , ,~ ~Ei 31 - AlectrlQ~iu.e,ctro1OphUfi- 87-B9 . .', . ': ". .' Page 31 - Ceb1diehtlrrs rlolaCewi - 89 , Page 35 - Liparis t!01:f-.e - 89 .Qhange: Page 11 - FmWulns parvipin¢.rl, add: Probable misidentification Page 20 - .BathopWuBt.lemin&, change to: .Mhgghilu§. llemipg+ Page 54 - Ji\mdJ11ui~~ add: Probable. misidentifioation Page 60 - Item. number 67, authOr should be .Hubbs, Clark TABLE OF CONTENTS INTRODUCTION 1 AREA OF COVERAGE 1 METHODS OF LITERATURE SEARCH 2 EXPLANATION OF CHECKLIST 2 ACKNOWLEDGEMENTS 4 TABLE 1 -

Life History Patterns and Biogeography: An
LIFE HISTORY PATTERNS AND Lynne R. Parenti2 BIOGEOGRAPHY: AN INTERPRETATION OF DIADROMY IN FISHES1 ABSTRACT Diadromy, broadly defined here as the regular movement between freshwater and marine habitats at some time during their lives, characterizes numerous fish and invertebrate taxa. Explanations for the evolution of diadromy have focused on ecological requirements of individual taxa, rarely reflecting a comparative, phylogenetic component. When incorporated into phylogenetic studies, center of origin hypotheses have been used to infer dispersal routes. The occurrence and distribution of diadromy throughout fish (aquatic non-tetrapod vertebrate) phylogeny are used here to interpret the evolution of this life history pattern and demonstrate the relationship between life history and ecology in cladistic biogeography. Cladistic biogeography has been mischaracterized as rejecting ecology. On the contrary, cladistic biogeography has been explicit in interpreting ecology or life history patterns within the broader framework of phylogenetic patterns. Today, in inferred ancient life history patterns, such as diadromy, we see remnants of previously broader distribution patterns, such as antitropicality or bipolarity, that spanned both marine and freshwater habitats. Biogeographic regions that span ocean basins and incorporate ocean margins better explain the relationship among diadromy, its evolution, and its distribution than do biogeographic regions centered on continents. Key words: Antitropical distributions, biogeography, diadromy, eels,