Journal of Experimental Biology and Agricultural Sciences FORENSIC
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
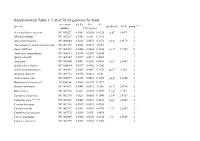
Supplementary Table 1: List of 76 Mt Genomes for Birds
Supplementary Table 1: List of 76 mt genomes for birds. accession Ka/Ks Ka Ks species speed(m/s) Ln S group * * number 13 mt genes Acrocephalus scirpaceus NC 010227 0.0464 0.0288 0.6228 6.6d 1.8871 2 Alectura lathami NC 007227 0.0343 0.032 0.9311 2 Anas platyrhynchos NC 009684 0.0252 0.0073 0.2873 18.5a 2.9178 2 Anomalopteryx didiformis (die out) NC 002779 0.0508 0.0027 0.054 1 Anser albifrons NC 004539 0.0468 0.0088 0.1856 16.1a 2.7788 2 Anseranas semipalmata NC 005933 0.0364 0.0207 0.5628 2 Apteryx haastii NC 002782 0.0599 0.0273 0.4561 1 Apus apus NC 008540 0.0401 0.0241 0.6431 10.6a 2.3609 2 Archilochus colubris NC 010094 0.0397 0.0382 0.9242 2 Ardea novaehollandiae NC 008551 0.0423 0.0067 0.1552 10.2c# 2.322 2 Arenaria interpres NC 003712 0.0378 0.0213 0.581 2 Aythya americana NC 000877 0.0309 0.0092 0.2987 24.4b 3.1946 2 Bambusicola thoracica* EU165706 0.0503 0.0144 0.2872 1 Branta canadensis NC 007011 0.0444 0.0092 0.206 16.7a 2.8154 2 Buteo buteo NC 003128 0.049 0.0167 0.3337 11.6a 2.451 2 Casuarius casuarius NC 002778 0.028 0.0085 0.3048 13.9f 2.6319 2 Cathartes aura * * * * NC 007628 0.0468 0.0239 0.4676 10.6c 2.3609 2 Ciconia boyciana NC 002196 0.0539 0.0031 0.0563 2 Ciconia ciconia NC 002197 0.0501 0.0029 0.0572 9.1d 2.2083 2 Cnemotriccus fuscatus NC 007975 0.0369 0.038 1.0476 2 Corvus frugilegus NC 002069 0.0428 0.0298 0.6526 13a 2.5649 2 Coturnix chinensis NC 004575 0.0325 0.0122 0.3762 2 Coturnix japonica NC 003408 0.0432 0.0128 0.2962 2 Cygnus columbianus NC 007691 0.0327 0.0089 0.2702 18.5a 2.9178 2 Dinornis giganteus -

Supporting Information
Supporting Information Bender et al. 10.1073/pnas.0802779105 Table S1. Species names and calculated numeric values pertaining to Fig. 1 Mitochondrially Nuclear encoded Nuclear encoded Species name encoded methionine, % methionine, % methionine, corrected, % Animals Primates Cebus albifrons 6.76 Cercopithecus aethiops 6.10 Colobus guereza 6.73 Gorilla gorilla 5.46 Homo sapiens 5.49 2.14 2.64 Hylobates lar 5.33 Lemur catta 6.50 Macaca mulatta 5.96 Macaca sylvanus 5.96 Nycticebus coucang 6.07 Pan paniscus 5.46 Pan troglodytes 5.38 Papio hamadryas 5.99 Pongo pygmaeus 5.04 Tarsius bancanus 6.74 Trachypithecus obscurus 6.18 Other mammals Acinonyx jubatus 6.86 Artibeus jamaicensis 6.43 Balaena mysticetus 6.04 Balaenoptera acutorostrata 6.17 Balaenoptera borealis 5.96 Balaenoptera musculus 5.78 Balaenoptera physalus 6.02 Berardius bairdii 6.01 Bos grunniens 7.04 Bos taurus 6.88 2.26 2.70 Canis familiaris 6.54 Capra hircus 6.84 Cavia porcellus 6.41 Ceratotherium simum 6.06 Choloepus didactylus 6.23 Crocidura russula 6.17 Dasypus novemcinctus 7.05 Didelphis virginiana 6.79 Dugong dugon 6.15 Echinops telfairi 6.32 Echinosorex gymnura 6.86 Elephas maximus 6.84 Equus asinus 6.01 Equus caballus 6.07 Erinaceus europaeus 7.34 Eschrichtius robustus 6.15 Eumetopias jubatus 6.77 Felis catus 6.62 Halichoerus grypus 6.46 Hemiechinus auritus 6.83 Herpestes javanicus 6.70 Hippopotamus amphibius 6.50 Hyperoodon ampullatus 6.04 Inia geoffrensis 6.20 Bender et al. www.pnas.org/cgi/content/short/0802779105 1of10 Mitochondrially Nuclear encoded Nuclear encoded Species -

2020 AAZV Proceedings.Pdf
PROCEEDINGS 2020 52ND AMERICAN ASSOCIATION OF ZOO VETERINARIANS Annual Conference 20 September - 24 September 2020 CHARLOTTE KIRK BAER PROCEEDINGS EDITOR CONTINUING EDUCATION Continuing education sponsored by the American College of Zoological Medicine. DISCLAIMER The information appearing in this publication comes exclusively from the authors and contributors identified in each manuscript. The techniques and procedures presented reflect the individual knowledge, experience, and personal views of the authors and contributors. The information presented does not incorporate all known techniques and procedures and is not exclusive. Other procedures, techniques, and technology might also be available. Any questions or requests for additional information concerning any of the manuscripts should be addressed directly to the authors. The sponsoring associations of this conference and resulting publication have not undertaken direct research or formal review to verify the information contained in this publication. Opinions expressed in this publication are those of the authors and contributors and do not necessarily reflect the views of the host associations. The associations are not responsible for errors or for opinions expressed in this publication. The host associations expressly disclaim any warranties or guarantees, expressed or implied, and shall not be liable for damages of any kind in connection with the material, information, techniques, or procedures set forth in this publication. AMERICAN ASSOCIATION OF ZOO VETERINARIANS “Dedicated to wildlife health and conservation” 581705 White Oak Road Yulee, Florida, 32097 904-225-3275 Fax 904-225-3289 Dear Friends and Colleagues, Welcome to our first-ever virtual AAZV Annual Conference! My deepest thanks to the AAZV Scientific Program Committee (SPC) and our other standing Committees for the work they have done to bring us to this point. -

Musica Poetica in Sixteenth-Century Reformation Germany
Musica Poetica in Sixteenth-Century Reformation Germany WONG,'Heleri]<:in Hoi .' ,. :,... ... ",-j." ... _ t.,~ . t " A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Mas'ter of Philosophy In Music The Chinese University of Hong Kong December 2009 Thesis Committee Professor OLSON Greta Jean (Chair) Professor Michael Edward MCCLELLAN (Thesis Supervisor) Professor Victor Amaro VICENTE (Committee Member) Professor MCKINNEY Timothy (External Examiner) Abstract of Thesis Entitled: Musica Poetica in Sixteenth-Century Reformation Germany Submitted by WONG, Helen Kin Hoi for the Degree of Master of Philosophy in Music at The Chinese University of Hong Kong in December 2009 ABSTRACT Musica poetica is a branch of music theory developed by German pedagogues of the Reformation era. It is the discipline within music composition that was grounded on the powerful relationships between music and text. The term musica poetica was first coined by the Lutheran musician/teacher Nicolaus Listenius in 1533 to distinguish it from musica theorica (the study of music as a mathematical science) and musica practica (applied theory dealing with aspects of performance), the two disciplines continuing from the Medieval education curriculum. By the middle of the sixteenth century, musica poetica began to be firmly established, alongside musica theorica and musica practica, as an independent branch of composition instruction, and was taught in the Latin schools of Lutheran Germany. As the treatises on musica poetica convey, the teaching of musica poetica was modelled on pedagogical principles of rhetoric that were taught in the humanistic curriculum of the Latin schools. In defining compositional procedures in relation to text-setting, these treatises borrowed or emulated terminologies from the discipline of classical rhetoric, and treated a musical composition as a work of oration, with an aesthetic aim of producing a work that could instruct, move and delight (docere, movere, delectare) the auditor. -

A Spiral Race Game from 17Th Century France
The Game of the Sphere or of the Universe | a Spiral Race Game from 17th century France Adrian Seville Emeritus Professor, City University, London Abstract Simple race games, played with dice and without choice of move, are known from antiquity. In the late 16th century, specific examples of this class of game emerged from Italy and spread rapidly into other countries of Europe. Pre-eminent was the Game of the Goose, which spawned thousands of variants over the succeeding centuries to the present day, including educational, polemical and promotional vari- ants.1 The educational variants began as a French invention of the 17th century, the earliest of known date being a game to teach Geography, the Jeu du Monde by Pierre Duval, published in 1645. By the end of the century, games designed to teach several of the other accomplishments required of the noble cadet class had been developed: History, the Arts of War, and Heraldry being notable among them. A remarkable example of a game within this class is the astronom- ical game, Le Jeu de la Sphere ou de l'Univers selon Tycho Brahe, published in 1661 by E(s)tienne Vouillemont in Paris. The present pa- per analyses this game in detail, showing how it combines four kinds of knowledge systems: natural philosophy, based on the Ptolemaic sphere; biblical knowledge; astrology, with planetary and zodiacal influences; and classical knowledge embodied in the names of the constellations. The game not only presents all four on an equal footing but also ex- plores links between them, indicating some acceptance of an overall knowledge-system. -

Constellations Concept Booklet Dan Lovallo 1 Constellations Algorithm A1) Selection of Constellation in Relation to Location: E.G
constellations concept booklet dan lovallo 1 constellations algorithm a1) Selection of constellation in relation to location: e.g. - Select constellations between Ecliptic and Celestial Equator - Number of constellation selection: 4 a2) Extraction of constellation in accordance to original default orientations CANCER a3) Extraction of constellation into vectorial points a1 a2 a3 b1) Overlapping of 4 sets of constellation points above one another. Sequence dependent on Constellation chart. b2) Vertical distances between each layer of constellation points relates to the relative real scale positions of those constellations. b3) Connection of points through single polyline to define total sequencing of constellation points. b1 b2 b3 c1) Panelling in accordance to the connected lines. Each panel is triangulated, governed by the closest two lines which form the edges of the panel. c2) Removal of panel based on random algorithm, which simulates the random connectivity of the constellations’ relative geographical locations. c3) Orientation of final output - select horizontal or vertical. c1 c2 c3 2 night sky photograph 3 visible stars chart 4 official constellations chart 5 chinese constellations chart Dunhuang Star map is one of the first known graphical representation of stars from ancient Chinese astronomy, dated to the Tang Dynasty (618–907) 6 chinese constellations chart 28 Chinese Constellations (asterisms) 7 chinese constellations chart 28 Chinese Constellations (asterisms) 8 egyptian constellations chart the zodiacal and para-zodiacal -

The Valley Skywatcher Official Publication of the Chagrin Valley Astronomical Society PO Box 11, Chagrin Falls, OH 44022 Founded 1963
The Valley Skywatcher Official Publication of the Chagrin Valley Astronomical Society PO Box 11, Chagrin Falls, OH 44022 www.chagrinvalleyastronomy.org Founded 1963 C ONTENTS O FFICERS F OR 2015 Articles President Marty Mullet Bruce Krobusek’s Vice President Ian Cooper Greatest Observing Project (So Far) 2 Treasurer Steve Fishman Regular Features Secretary Christina Gibbons Astrophotography 5 Directors of Observations Steve Kainec President’s Corner 6 Observatory Director Ken Fisher Observer’s Log 7 Constellation Quiz 9 Historian Dan Rothstein Notes & News 11 Editor Ron Baker Reflections 12 Solar activity recorded with a Coronado PST with Double Stack Filter on April 25, 2015. Image by Bruce Krobusek The Valley Skywatcher • Summer 2015 • Volume 52-3 • Page 1 Bruce Krobusek’s Greatest Observing Project (So Far) By Tony Mallama For those who may not know Bruce Krobusek or remember him from the club’s 50-year anniversary celebration, allow me to make a brief introduction. Bruce was president of the CVAS during the 1970s. Some say that he held the club together when it would have otherwise dissolved and Bruce received the club’s Backbone award for his efforts. His support for the CVAS has continued throughout the years in diverse ways including his editorship of the club’s history We Observe, his generous donation to fund the automation of the North Observatory and his submittal of articles and pictures to the Skywatcher. Bruce Krobusek in the 1970s with his 10-inch Deluxe Cave Astrola telescope in its fold-out-roof observatory. Bruce has always been an avid observer and, in the great tradition of amateur astronomers, has concentrated on re- cording observations that are useful to the science of astronomy. -

Star Tales 2018
CHAPTER ONE Stars and storytellers VERY NIGHT a pageant of Greek mythology circles overhead. Perseus flies to the rescue of Andromeda, Orion faces the charge of the snorting Ebull, Boötes herds the bears around the pole, and the ship of the Argo - nauts sails in search of the golden fleece. These legends, along with many others, are depicted in the star patterns that astronomers term constellations. Constellations are the invention of human imagination, not of nature. They are an expression of the human desire to impress its own order upon the appar- ent chaos of the night sky. For navigators beyond sight of land or for travellers in the trackless desert who wanted signposts, for farmers who wanted a calendar and for shepherds who wanted a nightly clock, the division of the sky into recognizable star groupings had practical purposes. But perhaps the earliest motivation was to humanize the forbidding blackness of night. Newcomers to astronomy are soon disappointed to find that the great majority of constellations bear little, if any, resemblance to the figures whose names they carry; but to expect such a resemblance is to misunderstand their true meaning. The constellation figures are not intended to be taken literally. Rather, they are symbolic, a celestial allegory. The night sky was a screen on which human imaginationSAMPLE could project the deeds and personifications of deities, sacred animals, and moral tales. It was a picture book in the days before writing. Each evening the stars emerge like magic spirits as the Sun descends to its nocturnal lair. Modern science has told us that those twinkling points scattered across the sky are actually glowing balls of gas similar to our own Sun, immensely far away. -

194 9 Ce Le B Rating 65 Ye Ars O F Br Inging As Tr on Omy T O No Rth Te X
1949 Celebrating 65 Years of Bringing Astronomy to North Texas 2014 Contact information: Inside this issue: Info Officer (General Info) – [email protected] Website Administrator – [email protected] Page Postal Address: October Club Calendar 3 Fort Worth Astronomical Society Celestial Events 4 c/o Matt McCullar 5801 Trail Lake Drive Sky Chart 5 Fort Worth, TX 76133 Web Site: http://www.fortworthastro.org Moon Phase Calendar 6 Facebook: http://tinyurl.com/3eutb22 Lunar Occultations/Conj 7 Twitter: http://twitter.com/ftwastro Yahoo! eGroup (members only): http://tinyurl.com/7qu5vkn Mars/Minor Planets Charts 8 Officers (2014-2015): Planet Vis & ISS Passes 9 President – Bruce Cowles, [email protected] Vice President – Russ Boatwright, [email protected] Young Astronomer News 10 Sec/Tres – Michelle Theisen, [email protected] Cloudy Night Library 11 Board Members: CSAC Invitation 13 2014-2016 Mike Langohr Seeing Spectrum 15 Tree Oppermann Lunar Eclipse Chart 17 2013-2015 18 Bill Nichols Solar Eclipse Chart Jim Craft Cover Photo: Monthly AL Observing Club 19 FWAS members attending our Sep- Constellation of the Month 20 tember 2014 star party/picnic at our Constellation Mythology 21 dark site in Montague County. Posing in front of our recently acquired 19- 22 Prior Club Meeting Minutes inch equatorial Newtonian reflector General Club Information 23 telescope. That’s A Fact 23 Observing Site Reminders: 23 Be careful with fire, mind all local burn bans! Full Moon Name Dark Site Usage Requirements (ALL MEMBERS): FWAS Foto Files 24 Maintain Dark-Sky Etiquette (http://tinyurl.com/75hjajy) Turn out your headlights at the gate! Sign the logbook (in camo-painted storage shed. -

Evolutionary Ecology of Giraffes (Giraffa Camelopardalis)
Evolutionary Ecology of Giraffes (Giraffa camelopardalis ) in Etosha National Park, Namibia Rachel Brand BSc Hons. Zoology A thesis submitted for the degree of Doctor of Philosophy (Ph.D) at Newcastle University School of Clinical Medical Sciences, Medical Faculty, NE2 4HH April 2007 2 ABSTRACT The giraffe ( Giraffa camelopardalis ) occupies a variety of habitats across sub- Saharan Africa. It is characterised by a loose social organisation, and a dominance- driven polygynous mating system. This project sought to explain biogeographic and inter-sexual variation in pelage colouration in the context of natural and sexual selection. I also sought to test the hypothesis that in a semi-arid environment, limited resources (food and water) would predictably concentrate females, increasing the potential for dominant males to monopolise matings. I analysed photos from across Africa, and reveal that where yearly bright sunshine is greater, female giraffe in particular tend to be lighter, resulting in sexual dichromatism in high insolarity locations. I hypothesised that dark pelage colour is maintained in males through sexual selection for a costly status signal. Field work was carried out in Etosha NP, Namibia. Using photographic records, I identified 431 individual giraffe. I surveyed the study area regularly and collected data on group composition and behaviour upon locating giraffe. I carried out focal watches, and recorded all observations of agonistic and mating behaviour. Darker males tended to be older and more dominant than lighter males, associated less with females, but had greater success in courting females. Food and water affected female movements on both a spatial and temporal scale. At waterholes, encounter rates were increased and consequently mating and agonistic interactions more frequent. -

Animals in “Light, Energy, and the EM Spectrum” Comic
Deborah Scherrer Stanford Solar Center Animals in “Light, Energy, and the EM Spectrum” Comic Found on which Species Information page? 22 The Bald Eagle has been the national emblem of the Bald Eagle United States since 1782 and a spiritual symbol for native people for far longer than that. These regal birds aren’t really bald, but their white-feathered heads gleam in contrast to their chocolate-brown body and wings. Look for the constellation Aquila to see an eagle in the sky. The stars are actually full of birds! Can you find Apus (the Bird of Paradise), Columba (the dove), Corvus (the crow), Cygnus (the swan), Grus (the crane), Pavo (the peacock), Phoenix (the mythical phoenix), and Tucana (the toucan)? Some birds navigate by the Sun, some by the stars, and others by the Earth’s magnetic field. http://www.birds.cornell.edu/allaboutbirds/studying/migr ation/navigation 3, 4, 12, Monarch butterflies are amongst the most beautiful of 22 Butterfly all butterflies and considered the “king” of the butterflies, hence the name “monarch”. These butterflies undertake a unique and miraculous migration Monarch every year. After wintering in Mexico or Pacific Grove, California, they begin a summer migration of 2500 miles. 2 additional generations are born, mate, lay eggs, and die during the migration, with a 4th generation hatching in Canada. This special 4th generation will spend the summer in Canada, then return to Mexico in the fall, flying the entire 2500 miles themselves. There Swallowtail they will winter-over, mate, lay eggs, and die. Their eggs will begin a new migration at the beginning of the next summer. -

NATURE in FOCUS Conservation Council of Europe
bulletin of the european information centre for nature NATURE IN FOCUS conservation council of europe I î> wildlife in the arctic ~\ " ■N d r " IT -J r e u ro p e a n — u [Ï information min c e n tre L h J V. J for Urniü. n atu re number 11 winter 1971 -72 conservation NATURE IN FOCUS Everywhere the ancient cities and acter; the building of roads to divert picturesque villages, which we love traffic away from areas of architec Editorial The Rt Hon Duncan Sandys, MP 1 and which tourists come from afar to tural interest or to by-pass charming admire, are being progressively de villages; the elimination of car parking molished or mutilated for commercial in fine squares and streets; the crea Transfrontier natural parks Dr Hertha Firnberg 2 gain or the convenience of motor tion of pedestrian precincts; the re traffic. If this process is allowed to moval of unsightly outdoor advertising, Nature and history: a common heritage for continue unchecked, Europe's distinc overhead wires and other ugliness; tive character will soon be totally de the careful planning of new develop Conservation Bernard Champigneulle 5 stroyed. All that will be left will be a ment in areas of scenic beauty on the few isolated monuments retained as coast, in the countryside or in the Controlling traffic in wild animals Moira a g wariand 7 lifeless museums in the midst of a mountains; the introduction of more jungle of ferro concrete and tarmac. trees and grass in towns and villages; Whether it be the historic centre of a the elimination of dirt, decay and un