Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
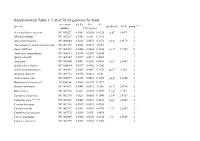
Supplementary Table 1: List of 76 Mt Genomes for Birds
Supplementary Table 1: List of 76 mt genomes for birds. accession Ka/Ks Ka Ks species speed(m/s) Ln S group * * number 13 mt genes Acrocephalus scirpaceus NC 010227 0.0464 0.0288 0.6228 6.6d 1.8871 2 Alectura lathami NC 007227 0.0343 0.032 0.9311 2 Anas platyrhynchos NC 009684 0.0252 0.0073 0.2873 18.5a 2.9178 2 Anomalopteryx didiformis (die out) NC 002779 0.0508 0.0027 0.054 1 Anser albifrons NC 004539 0.0468 0.0088 0.1856 16.1a 2.7788 2 Anseranas semipalmata NC 005933 0.0364 0.0207 0.5628 2 Apteryx haastii NC 002782 0.0599 0.0273 0.4561 1 Apus apus NC 008540 0.0401 0.0241 0.6431 10.6a 2.3609 2 Archilochus colubris NC 010094 0.0397 0.0382 0.9242 2 Ardea novaehollandiae NC 008551 0.0423 0.0067 0.1552 10.2c# 2.322 2 Arenaria interpres NC 003712 0.0378 0.0213 0.581 2 Aythya americana NC 000877 0.0309 0.0092 0.2987 24.4b 3.1946 2 Bambusicola thoracica* EU165706 0.0503 0.0144 0.2872 1 Branta canadensis NC 007011 0.0444 0.0092 0.206 16.7a 2.8154 2 Buteo buteo NC 003128 0.049 0.0167 0.3337 11.6a 2.451 2 Casuarius casuarius NC 002778 0.028 0.0085 0.3048 13.9f 2.6319 2 Cathartes aura * * * * NC 007628 0.0468 0.0239 0.4676 10.6c 2.3609 2 Ciconia boyciana NC 002196 0.0539 0.0031 0.0563 2 Ciconia ciconia NC 002197 0.0501 0.0029 0.0572 9.1d 2.2083 2 Cnemotriccus fuscatus NC 007975 0.0369 0.038 1.0476 2 Corvus frugilegus NC 002069 0.0428 0.0298 0.6526 13a 2.5649 2 Coturnix chinensis NC 004575 0.0325 0.0122 0.3762 2 Coturnix japonica NC 003408 0.0432 0.0128 0.2962 2 Cygnus columbianus NC 007691 0.0327 0.0089 0.2702 18.5a 2.9178 2 Dinornis giganteus -

A Higher-Level Phylogenetic Classification of the Fungi
mycological research 111 (2007) 509–547 available at www.sciencedirect.com journal homepage: www.elsevier.com/locate/mycres A higher-level phylogenetic classification of the Fungi David S. HIBBETTa,*, Manfred BINDERa, Joseph F. BISCHOFFb, Meredith BLACKWELLc, Paul F. CANNONd, Ove E. ERIKSSONe, Sabine HUHNDORFf, Timothy JAMESg, Paul M. KIRKd, Robert LU¨ CKINGf, H. THORSTEN LUMBSCHf, Franc¸ois LUTZONIg, P. Brandon MATHENYa, David J. MCLAUGHLINh, Martha J. POWELLi, Scott REDHEAD j, Conrad L. SCHOCHk, Joseph W. SPATAFORAk, Joost A. STALPERSl, Rytas VILGALYSg, M. Catherine AIMEm, Andre´ APTROOTn, Robert BAUERo, Dominik BEGEROWp, Gerald L. BENNYq, Lisa A. CASTLEBURYm, Pedro W. CROUSl, Yu-Cheng DAIr, Walter GAMSl, David M. GEISERs, Gareth W. GRIFFITHt,Ce´cile GUEIDANg, David L. HAWKSWORTHu, Geir HESTMARKv, Kentaro HOSAKAw, Richard A. HUMBERx, Kevin D. HYDEy, Joseph E. IRONSIDEt, Urmas KO˜ LJALGz, Cletus P. KURTZMANaa, Karl-Henrik LARSSONab, Robert LICHTWARDTac, Joyce LONGCOREad, Jolanta MIA˛ DLIKOWSKAg, Andrew MILLERae, Jean-Marc MONCALVOaf, Sharon MOZLEY-STANDRIDGEag, Franz OBERWINKLERo, Erast PARMASTOah, Vale´rie REEBg, Jack D. ROGERSai, Claude ROUXaj, Leif RYVARDENak, Jose´ Paulo SAMPAIOal, Arthur SCHU¨ ßLERam, Junta SUGIYAMAan, R. Greg THORNao, Leif TIBELLap, Wendy A. UNTEREINERaq, Christopher WALKERar, Zheng WANGa, Alex WEIRas, Michael WEISSo, Merlin M. WHITEat, Katarina WINKAe, Yi-Jian YAOau, Ning ZHANGav aBiology Department, Clark University, Worcester, MA 01610, USA bNational Library of Medicine, National Center for Biotechnology Information, -

SYSTEMATICS of DIVISION ASCOMYCOTA 1 Group
References: Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008. Dictionary of the Fungi (10th ed.). Wallingford, UK: CABI. Webster, J., & Weber, R. (2007). Introduction to fungi. Cambridge, UK: Cambridge University Press. Url1.: https://en.wikipedia.org/wiki/Ascomycota SYSTEMATICS OF DIVISION ASCOMYCOTA 1 Group: Plectomycetes Plectomycetes is an artificial group of Ascomycota and it originally contained all Ascomycete fungi which produce their asci within a cleistothecium. Plectomycetes can be defined by the following set of characters; Cleistothecium or gymnothecium is usually present, ascogenous hyphae are usually not conspicuous, asci are scattered throughout the cleistothecium, asci are mostly globose and thin-walled, and the ascospores are released passively after disintegration of the ascus wall, not by active discharge, ascospores are small, unicellular and usually spherical or ovoid, conidia are commonly produced from phialides or as arthroconidia. Class: Eurotiomycetes Most members of the class produce an enclosed structure cleistothecium within which they produce their spores. It contains 10 order, 27 families 280 genus and about 3400 species. Order: Onygenales Onygenales members are able to digest keratin and because of this have become dominant organisms in environments where keratin is available. The most members have colorless cleistothecia and ascospores. The spherical to egg-shaped asci are always uniformly packed in the centrum and may be dispersed among hyphal elements. The ascospores are always single-celled (example: Chrysosporium, Microsporum and Trichophyton). Order: Eurotiales Most members of the order have phialidic asexual stages belonging to the genera Aspergillus and Penicillium or, less commonly, to Paecilomyces or even simpler types. Rarely there is no anamorph at all. -

Phylogenomic and Comparative Analysis of the Distribution And
www.nature.com/scientificreports OPEN Phylogenomic and comparative analysis of the distribution and regulatory patterns of TPP Received: 25 August 2017 Accepted: 20 March 2018 riboswitches in fungi Published: xx xx xxxx Sumit Mukherjee1, Matan Drory Retwitzer2, Danny Barash2 & Supratim Sengupta 1 Riboswitches are metabolite or ion sensing cis-regulatory elements that regulate the expression of the associated genes involved in biosynthesis or transport of the corresponding metabolite. Among the nearly 40 diferent classes of riboswitches discovered in bacteria so far, only the TPP riboswitch has also been found in algae, plants, and in fungi where their presence has been experimentally validated in a few instances. We analyzed all the available complete fungal and related genomes and identifed TPP riboswitch-based regulation systems in 138 fungi and 15 oomycetes. We fnd that TPP riboswitches are most abundant in Ascomycota and Basidiomycota where they regulate TPP biosynthesis and/ or transporter genes. Many of these transporter genes were found to contain conserved domains consistent with nucleoside, urea and amino acid transporter gene families. The genomic location of TPP riboswitches when correlated with the intron structure of the regulated genes enabled prediction of the precise regulation mechanism employed by each riboswitch. Our comprehensive analysis of TPP riboswitches in fungi provides insights about the phylogenomic distribution, regulatory patterns and functioning mechanisms of TPP riboswitches across diverse fungal species and provides a useful resource that will enhance the understanding of RNA-based gene regulation in eukaryotes. Riboswitches are conserved non-coding structural RNA sensors that bind specifc metabolites and regulate gene expression1–4. A riboswitch is mainly composed of two domains; a highly conserved aptamer domain, which is responsible for binding of metabolites, and the expression platform that changes conformation on metabolite binding and regulates the expression of associated genes2. -

Fungal Genome Initiative
Fungal Genome Initiative A White Paper for Fungal Genomics July 10, 2004 Submitted on behalf of: The Fungal Genome Initiative Steering Committee Corresponding author: Bruce Birren The Broad Institute of Harvard and MIT 320 Charles Street, Cambridge, MA 02141 USA. Phone: 617-258-0900. E-mail: [email protected]. Summary Since 2001 fungal scientists and members of the Broad Institute have been pursuing a comprehensive plan to apply comparative sequence analysis to study the evolution of eukaryotic cellular processes and the molecular basis for fungal pathogenesis. In this white paper we seek approval to sequence four fungi to continue these efforts: • Schizosaccharomyces octosporus and Schizosaccharomyces japonicus — to improve the annotation of the S. pombe genome sequence. • Trichophyton rubrum — the cause of athlete’s foot and the first organism to be sequenced in the class of fungi that adhere to human skin. • Batrachochytrium dendrobatidis — the causal organism in the recent dramatic decline of global amphibian populations. The sequences of these four genomes, totaling 88 Mb, will both directly contribute to the study of key fungal pathogens and advance the utility of S. pombe, a widely used model for molecular and cellular research. Background and rationale Although S. cerevisiae, S. pombe and several filamentous fungi are well-established research organisms, the vast majority of fungi remain very poorly understood. The fact that over 800 million years (myr) has elapsed since the divergence from the common ancestor of the fungal kingdom (Figure1) means that these few well-studied organisms do not adequately represent the diversity found within the many fungi that play critical roles in human health and industry. -

MATING SYSTEM and MITOCHONDRIAL INHERITANCE in a BASIDIOMYCETE YEAST, CRYPTOCOCCUS NEOFORMANS Phd Thesis-Zhun Yan__ Mcmaster University- Biology
PhD Thesis-Zhun Yan __ McMaster University- Biology MATING SYSTEM AND MITOCHONDRIAL INHERITANCE IN A BASIDIOMYCETE YEAST, CRYPTOCOCCUS NEOFORMANS PhD Thesis-Zhun Yan__ McMaster University- Biology MA1['ING SYSTEM AND MITOCHONDRIAL INHEJliTANCE IN A BASIDIOMYCETE YEAST, CRYPTOCOCCUS NEOFORMANS By ZHUNYAN A Thesis Submitted to the School of Graduate Studies in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy McMaster University Copyright by Zhun Yan, March 2006 PhD Thesis-Zhun Y an__ McMaster University- Biology Doctor of Philowphy (2006) McMaster University (Biology) Hamilton, Ontario TITLE: Mating system and mitochondrial inheritance in a basidiomycete yeast, Cryptococcus neoformans AUTHOR: Zhun Yan, B.Sc., M.Sc. SUPERVISOR: :Dr. Jianping Xu NUMBER OF PAGES: xii, 202 ii PhD Thesis-Zhun Yan__ McMaster University- Biology ABSTRACT In the majority of sexual eukaryotes, mitochondria are inherited predominantly from a single, usually the female, parent Like the majority of higher plants and animals, the pathogenic yeas1 Cryptococcus neoformans has two mating types (sexes), however, these two sexes are morphologically similar. In this study, I examined the distribution of the mating tyres and how mating types influence the inheritance of mitochondria in C. neoformans. My survey of mating type alleles in 358 isolates collected from four geographic areas in the US showed a biased distribution of mating type alleles with most isolates containing mating type a alleles. To characterize the role of mating type locus on mitochondrial inheritance, I constructed two pairs of congenic strains that differed only at the mitochondrial genome and mating type locus. Mating between these two pairs of stra ms demonstrated that uniparental inheritance in C. -

2020 AAZV Proceedings.Pdf
PROCEEDINGS 2020 52ND AMERICAN ASSOCIATION OF ZOO VETERINARIANS Annual Conference 20 September - 24 September 2020 CHARLOTTE KIRK BAER PROCEEDINGS EDITOR CONTINUING EDUCATION Continuing education sponsored by the American College of Zoological Medicine. DISCLAIMER The information appearing in this publication comes exclusively from the authors and contributors identified in each manuscript. The techniques and procedures presented reflect the individual knowledge, experience, and personal views of the authors and contributors. The information presented does not incorporate all known techniques and procedures and is not exclusive. Other procedures, techniques, and technology might also be available. Any questions or requests for additional information concerning any of the manuscripts should be addressed directly to the authors. The sponsoring associations of this conference and resulting publication have not undertaken direct research or formal review to verify the information contained in this publication. Opinions expressed in this publication are those of the authors and contributors and do not necessarily reflect the views of the host associations. The associations are not responsible for errors or for opinions expressed in this publication. The host associations expressly disclaim any warranties or guarantees, expressed or implied, and shall not be liable for damages of any kind in connection with the material, information, techniques, or procedures set forth in this publication. AMERICAN ASSOCIATION OF ZOO VETERINARIANS “Dedicated to wildlife health and conservation” 581705 White Oak Road Yulee, Florida, 32097 904-225-3275 Fax 904-225-3289 Dear Friends and Colleagues, Welcome to our first-ever virtual AAZV Annual Conference! My deepest thanks to the AAZV Scientific Program Committee (SPC) and our other standing Committees for the work they have done to bring us to this point. -

Musica Poetica in Sixteenth-Century Reformation Germany
Musica Poetica in Sixteenth-Century Reformation Germany WONG,'Heleri]<:in Hoi .' ,. :,... ... ",-j." ... _ t.,~ . t " A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Mas'ter of Philosophy In Music The Chinese University of Hong Kong December 2009 Thesis Committee Professor OLSON Greta Jean (Chair) Professor Michael Edward MCCLELLAN (Thesis Supervisor) Professor Victor Amaro VICENTE (Committee Member) Professor MCKINNEY Timothy (External Examiner) Abstract of Thesis Entitled: Musica Poetica in Sixteenth-Century Reformation Germany Submitted by WONG, Helen Kin Hoi for the Degree of Master of Philosophy in Music at The Chinese University of Hong Kong in December 2009 ABSTRACT Musica poetica is a branch of music theory developed by German pedagogues of the Reformation era. It is the discipline within music composition that was grounded on the powerful relationships between music and text. The term musica poetica was first coined by the Lutheran musician/teacher Nicolaus Listenius in 1533 to distinguish it from musica theorica (the study of music as a mathematical science) and musica practica (applied theory dealing with aspects of performance), the two disciplines continuing from the Medieval education curriculum. By the middle of the sixteenth century, musica poetica began to be firmly established, alongside musica theorica and musica practica, as an independent branch of composition instruction, and was taught in the Latin schools of Lutheran Germany. As the treatises on musica poetica convey, the teaching of musica poetica was modelled on pedagogical principles of rhetoric that were taught in the humanistic curriculum of the Latin schools. In defining compositional procedures in relation to text-setting, these treatises borrowed or emulated terminologies from the discipline of classical rhetoric, and treated a musical composition as a work of oration, with an aesthetic aim of producing a work that could instruct, move and delight (docere, movere, delectare) the auditor. -

Division: Ascomycota Division Ascomycota Is the Largest Fungal
Division: Ascomycota Division Ascomycota is the largest fungal division which contains approximately 75% of all described fungi. The division includes 15 class, 68 order, 327 families, 6355 genera and approximately 64000 species. It is a morphologically diverse division which contains organisms from unicellular yeasts to complex cup fungi. Most of its members are terrestrial or parasitic. However, a few have adapted to marine or freshwater environments. Some of them form symbiotic associations with algae to form lichens. The division members, commonly known as the sac fungi, are characterized by the presence of a reproductive microscopic sexual structure called ascus in which ascospores are formed. Nuclear fusion and meiosis occur in the ascus and one round of mitosis typically follows meiosis to leave eight nuclei. Finally, eight ascospores take place. Ascospores are formed within the ascus by an enveloping membrane system, which packages each nucleus with its adjacent cytoplasm and provides the site for ascospore wall formation. Another unique character of the division (but not present in all ascomycetes) is the presence of Woronin bodies on each side of the septa separating the hyphal segments which control the septal pores. Like all fungi, The cell walls of the hyphae of Ascomycota are variably composed of chitin and β-glucans. The mycelia of the division usually consist of septate hyphae. Its septal walls have septal pores which provide cytoplasmic continuity throughout the individual hyphae. Under appropriate conditions, nuclei may also migrate between septal compartments through the septal pores. Asexual reproduction of Ascomycota is responsible for rapid reproduction. It takes places through vegetative reproductive spores called conidia but chlamydospores are also frequently produced. -

Evidence for Inter-Specific Recombination Among the Mitochondrial Genomes of Fusarium Species in the Gibberella Fujikuroi Comple
Fourie et al. BMC Genomics 2013, 14:605 http://www.biomedcentral.com/1471-2164/14/605 RESEARCH ARTICLE Open Access Evidence for inter-specific recombination among the mitochondrial genomes of Fusarium species in the Gibberella fujikuroi complex Gerda Fourie1*, Nicolaas A van der Merwe2, Brenda D Wingfield2, Mesfin Bogale2, Bettina Tudzynski3, Michael J Wingfield1 and Emma T Steenkamp1 Abstract Background: The availability of mitochondrial genomes has allowed for the resolution of numerous questions regarding the evolutionary history of fungi and other eukaryotes. In the Gibberella fujikuroi species complex, the exact relationships among the so-called “African”, “Asian” and “American” Clades remain largely unresolved, irrespective of the markers employed. In this study, we considered the feasibility of using mitochondrial genes to infer the phylogenetic relationships among Fusarium species in this complex. The mitochondrial genomes of representatives of the three Clades (Fusarium circinatum, F. verticillioides and F. fujikuroi) were characterized and we determined whether or not the mitochondrial genomes of these fungi have value in resolving the higher level evolutionary relationships in the complex. Results: Overall, the mitochondrial genomes of the three species displayed a high degree of synteny, with all the genes (protein coding genes, unique ORFs, ribosomal RNA and tRNA genes) in identical order and orientation, as well as introns that share similar positions within genes. The intergenic regions and introns generally contributed significantly to the size differences and diversity observed among these genomes. Phylogenetic analysis of the concatenated protein-coding dataset separated members of the Gibberella fujikuroi complex from other Fusarium species and suggested that F. fujikuroi (“Asian” Clade) is basal in the complex. -

The Abundance and Diversity of Fungi in a Hypersaline Microbial Mat from Guerrero Negro, Baja California, México
Journal of Fungi Article The Abundance and Diversity of Fungi in a Hypersaline Microbial Mat from Guerrero Negro, Baja California, México Paula Maza-Márquez 1,*, Michael D. Lee 1,2 and Brad M. Bebout 1 1 Exobiology Branch, NASA Ames Research Center, Moffett Field, CA 94035, USA; [email protected] (M.D.L.); [email protected] (B.M.B.) 2 Blue Marble Space Institute of Science, Seattle, WA 98104, USA * Correspondence: [email protected] Abstract: The abundance and diversity of fungi were evaluated in a hypersaline microbial mat from Guerrero Negro, México, using a combination of quantitative polymerase chain reaction (qPCR) amplification of domain-specific primers, and metagenomic sequencing. Seven different layers were analyzed in the mat (Layers 1–7) at single millimeter resolution (from the surface to 7 mm in depth). The number of copies of the 18S rRNA gene of fungi ranged between 106 and 107 copies per g mat, being two logarithmic units lower than of the 16S rRNA gene of bacteria. The abundance of 18S rRNA genes of fungi varied significantly among the layers with layers 2–5 mm from surface contained the highest numbers of copies. Fifty-six fungal taxa were identified by metagenomic sequencing, classified into three different phyla: Ascomycota, Basidiomycota and Microsporidia. The prevalent genera of fungi were Thermothelomyces, Pyricularia, Fusarium, Colletotrichum, Aspergillus, Botrytis, Candida and Neurospora. Genera of fungi identified in the mat were closely related to genera known to have saprotrophic and parasitic lifestyles, as well as genera related to human and plant pathogens and fungi able to perform denitrification. -

High-Level Classification of the Fungi and a Tool for Evolutionary Ecological Analyses
Fungal Diversity (2018) 90:135–159 https://doi.org/10.1007/s13225-018-0401-0 (0123456789().,-volV)(0123456789().,-volV) High-level classification of the Fungi and a tool for evolutionary ecological analyses 1,2,3 4 1,2 3,5 Leho Tedersoo • Santiago Sa´nchez-Ramı´rez • Urmas Ko˜ ljalg • Mohammad Bahram • 6 6,7 8 5 1 Markus Do¨ ring • Dmitry Schigel • Tom May • Martin Ryberg • Kessy Abarenkov Received: 22 February 2018 / Accepted: 1 May 2018 / Published online: 16 May 2018 Ó The Author(s) 2018 Abstract High-throughput sequencing studies generate vast amounts of taxonomic data. Evolutionary ecological hypotheses of the recovered taxa and Species Hypotheses are difficult to test due to problems with alignments and the lack of a phylogenetic backbone. We propose an updated phylum- and class-level fungal classification accounting for monophyly and divergence time so that the main taxonomic ranks are more informative. Based on phylogenies and divergence time estimates, we adopt phylum rank to Aphelidiomycota, Basidiobolomycota, Calcarisporiellomycota, Glomeromycota, Entomoph- thoromycota, Entorrhizomycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota and Olpidiomycota. We accept nine subkingdoms to accommodate these 18 phyla. We consider the kingdom Nucleariae (phyla Nuclearida and Fonticulida) as a sister group to the Fungi. We also introduce a perl script and a newick-formatted classification backbone for assigning Species Hypotheses into a hierarchical taxonomic framework, using this or any other classification system. We provide an example