Phage Display-Based Strategies for Cloning and Optimization of Monoclonal Antibodies Directed Against Human Pathogens
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

WO 2015/063180 Al 7 May 2015 (07.05.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/063180 Al 7 May 2015 (07.05.2015) P O P C T (51) International Patent Classification: (74) Agent: WILLIAMS, Rachel; Novozymes Biopharma UK A61K 39/00 (2006.01) C07K 16/00 (2006.01) Ltd., Castle Court, 59 Castle Boulevard, Nottingham Not A61K 39/395 (2006.01) tinghamshire NG7 1FD (GB). (21) International Application Number: (81) Designated States (unless otherwise indicated, for every PCT/EP2014/073261 kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (22) International Filing Date: BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, 29 October 2014 (29.10.2014) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (25) Filing Language: English HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (26) Publication Language: English MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, (30) Priority Data: PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, 13 190750.3 29 October 2013 (29. 10.2013) EP SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 14166865.7 2 May 2014 (02.05.2014) EP TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (71) Applicant: NOVOZYMES BIOPHARMA DK A S (84) Designated States (unless otherwise indicated, for every [DK/DK]; Krogshoejvej 36, DK-2880 Bagsvaerd (DK). -

Predictive QSAR Tools to Aid in Early Process Development of Monoclonal Antibodies
Predictive QSAR tools to aid in early process development of monoclonal antibodies John Micael Andreas Karlberg Published work submitted to Newcastle University for the degree of Doctor of Philosophy in the School of Engineering November 2019 Abstract Monoclonal antibodies (mAbs) have become one of the fastest growing markets for diagnostic and therapeutic treatments over the last 30 years with a global sales revenue around $89 billion reported in 2017. A popular framework widely used in pharmaceutical industries for designing manufacturing processes for mAbs is Quality by Design (QbD) due to providing a structured and systematic approach in investigation and screening process parameters that might influence the product quality. However, due to the large number of product quality attributes (CQAs) and process parameters that exist in an mAb process platform, extensive investigation is needed to characterise their impact on the product quality which makes the process development costly and time consuming. There is thus an urgent need for methods and tools that can be used for early risk-based selection of critical product properties and process factors to reduce the number of potential factors that have to be investigated, thereby aiding in speeding up the process development and reduce costs. In this study, a framework for predictive model development based on Quantitative Structure- Activity Relationship (QSAR) modelling was developed to link structural features and properties of mAbs to Hydrophobic Interaction Chromatography (HIC) retention times and expressed mAb yield from HEK cells. Model development was based on a structured approach for incremental model refinement and evaluation that aided in increasing model performance until becoming acceptable in accordance to the OECD guidelines for QSAR models. -
Inclusion and Exclusion Criteria for Each Key Question
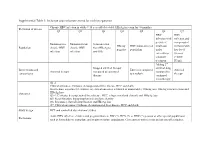
Supplemental Table 1: Inclusion and exclusion criteria for each key question Chronic HBV infection in adults ≥ 18 year old (detectable HBsAg in serum for >6 months) Definition of disease Q1 Q2 Q3 Q4 Q5 Q6 Q7 HBV HBV infection with infection and persistent compensated Immunoactive Immunotolerant Seroconverted HBeAg HBV mono-infected viral load cirrhosis with Population chronic HBV chronic HBV from HBeAg to negative population under low level infection infection anti-HBe entecavir or viremia tenofovir (<2000 treatment IU/ml) Adding 2nd Stopped antiviral therapy antiviral drug Interventions and Entecavir compared Antiviral Antiviral therapy compared to continued compared to comparisons to tenofovir therapy therapy continued monotherapy Q1-2: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Intermediate outcomes (if evidence on clinical outcomes is limited or unavailable): HBsAg loss, HBeAg seroconversion and Outcomes HBeAg loss Q3-4: Cirrhosis, decompensated liver disease, HCC, relapse (viral and clinical) and HBsAg loss Q5: Renal function, hypophosphatemia and bone density Q6: Resistance, flare/decompensation and HBeAg loss Q7: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Study design RCT and controlled observational studies Acute HBV infection, children and pregnant women, HIV (+), HCV (+) or HDV (+) persons or other special populations Exclusions such as hemodialysis, transplant, and treatment failure populations. Co treatment with steroids and uncontrolled studies. Supplemental Table 2: Detailed Search Strategy: Ovid Database(s): Embase 1988 to 2014 Week 37, Ovid MEDLINE(R) In-Process & Other Non- Indexed Citations and Ovid MEDLINE(R) 1946 to Present, EBM Reviews - Cochrane Central Register of Controlled Trials August 2014, EBM Reviews - Cochrane Database of Systematic Reviews 2005 to July 2014 Search Strategy: # Searches Results 1 exp Hepatitis B/dt 26410 ("hepatitis B" or "serum hepatitis" or "hippie hepatitis" or "injection hepatitis" or 2 178548 "hepatitis type B").mp. -

(12) United States Patent (10) Patent No.: US 9.468,689 B2 Zeng Et Al
USOO9468689B2 (12) United States Patent (10) Patent No.: US 9.468,689 B2 Zeng et al. (45) Date of Patent: *Oct. 18, 2016 (54) ULTRAFILTRATION CONCENTRATION OF (56) References Cited ALLOTYPE SELECTED ANTIBODES FOR SMALL-VOLUME ADMINISTRATION U.S. PATENT DOCUMENTS 5,429,746 A 7/1995 Shadle et al. (71) Applicant: Immunomedics, Inc., Morris Plains, NJ 5,789,554 A 8/1998 Leung et al. (US) 6,171,586 B1 1/2001 Lam et al. 6,187,287 B1 2/2001 Leung et al. (72) Inventors: Li Zeng, Edison, NJ (US); Rohini 6,252,055 B1 6/2001 Relton Mitra, Brigdewater, NJ (US); Edmund 6,676,924 B2 1/2004 Hansen et al. 6,870,034 B2 3/2005 Breece et al. A. Rossi, Woodland Park, NJ (US); 6,893,639 B2 5/2005 Levy et al. Hans J. Hansen, Picayune, MS (US); 6,991,790 B1 1/2006 Lam et al. David M. Goldenberg, Mendham, NJ 7,038,017 B2 5, 2006 Rinderknecht et al. (US) 7,074,403 B1 7/2006 Goldenberg et al. 7,109,304 B2 9, 2006 Hansen et al. 7,138,496 B2 11/2006 Hua et al. (73) Assignee: Immunomedics, Inc., Morris Plains, NJ 7,151,164 B2 * 12/2006 Hansen et al. ............. 530,387.3 (US) 7,238,785 B2 7/2007 Govindan et al. 7,251,164 B2 7/2007 Okhonin et al. (*) Notice: Subject to any disclaimer, the term of this 7.282,567 B2 10/2007 Goldenberg et al. patent is extended or adjusted under 35 7,300,655 B2 11/2007 Hansen et al. -

AHFS Pharmacologic-Therapeutic Classification System
AHFS Pharmacologic-Therapeutic Classification System Abacavir 48:24 - Mucolytic Agents - 382638 8:18.08.20 - HIV Nucleoside and Nucleotide Reverse Acitretin 84:92 - Skin and Mucous Membrane Agents, Abaloparatide 68:24.08 - Parathyroid Agents - 317036 Aclidinium Abatacept 12:08.08 - Antimuscarinics/Antispasmodics - 313022 92:36 - Disease-modifying Antirheumatic Drugs - Acrivastine 92:20 - Immunomodulatory Agents - 306003 4:08 - Second Generation Antihistamines - 394040 Abciximab 48:04.08 - Second Generation Antihistamines - 394040 20:12.18 - Platelet-aggregation Inhibitors - 395014 Acyclovir Abemaciclib 8:18.32 - Nucleosides and Nucleotides - 381045 10:00 - Antineoplastic Agents - 317058 84:04.06 - Antivirals - 381036 Abiraterone Adalimumab; -adaz 10:00 - Antineoplastic Agents - 311027 92:36 - Disease-modifying Antirheumatic Drugs - AbobotulinumtoxinA 56:92 - GI Drugs, Miscellaneous - 302046 92:20 - Immunomodulatory Agents - 302046 92:92 - Other Miscellaneous Therapeutic Agents - 12:20.92 - Skeletal Muscle Relaxants, Miscellaneous - Adapalene 84:92 - Skin and Mucous Membrane Agents, Acalabrutinib 10:00 - Antineoplastic Agents - 317059 Adefovir Acamprosate 8:18.32 - Nucleosides and Nucleotides - 302036 28:92 - Central Nervous System Agents, Adenosine 24:04.04.24 - Class IV Antiarrhythmics - 304010 Acarbose Adenovirus Vaccine Live Oral 68:20.02 - alpha-Glucosidase Inhibitors - 396015 80:12 - Vaccines - 315016 Acebutolol Ado-Trastuzumab 24:24 - beta-Adrenergic Blocking Agents - 387003 10:00 - Antineoplastic Agents - 313041 12:16.08.08 - Selective -

Anticuerpos Monoclonales: Desarrollo Físico Y Perspectivas Terapéuticas
NINA PATRICIA MACHADO ET. AL ARTÍCULO DE REVISIÓN Anticuerpos monoclonales: desarrollo físico y perspectivas terapéuticas Monoclonal antibodies: physical development and therapeutic perspectives NINA PATRICIA MACHADO1, GERMÁN ALBERTO TÉLLEZ2, JOHN CARLOS CASTAÑO2 Resumen Los anticuerpos monoclonales son glucoproteínas Abstract especializadas que hacen parte del sistema inmune, Monoclonal antibodies are specialized glucopro- producidas por las células B, con la capacidad de re- teins that belong to the immune system, produced conocer moléculas específicas (antígenos). Los anti- by the B cells which have the ability to recognize cuerpos monoclonales son herramientas esenciales other molecules (antigens). They are important en el ámbito clínico y biotecnológico, y han probado tools in clinical practice and biotechnology and have ser útiles en el diagnóstico y tratamiento de enfer- been useful in the diagnosis and treatment of medades infecciosas, inmunológicas y neoplá-sicas, infectious, inflammatory, immunological and así como también en el estudio de las interacciones neoplasic diseases, as well as in the study of the patógeno-hospedero y la marcación, detección y cuan- host/patogen interaction, and in the detection and tificación de diversas moléculas. quantification of diverse molecules. Actualmente, la incorporación de las técnicas The incorporation of molecular biology, de biología molecular e ingeniería genética y proteica proteic and genetic engineering have extended the han permitido ampliar el horizonte de la generación production -

WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C07D 519/00 (2006.01) A61P 39/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C07D 487/04 (2006.01) A61P 35/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 31/5517 (2006.01) A61P 37/00 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, A61K 47/48 (2006.01) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (21) International Application Number: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, PCT/IB2013/058229 SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (22) International Filing Date: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2 September 2013 (02.09.2013) ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: HANGZHOU DAC BIOTECH CO., LTD UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, [US/CN]; Room B2001-B2019, Building 2, No 452 Sixth TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, Street, Hangzhou Economy Development Area, Hangzhou EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, City, Zhejiang 310018 (CN). -

1 Therapeutic Antibodies – from Past to Future Stefan Dubel¨ and Janice M
1 1 Therapeutic Antibodies – from Past to Future Stefan Dubel¨ and Janice M. Reichert 1.1 An Exciting Start – and a Long Trek In the late nineteenth century, the German army doctor Emil von Behring (1854–1917), later the first Nobel Laureate for Medicine, pioneered the thera- peutic application of antibodies. He used blood serum for the treatment of tetanus and diphtheria (‘‘Blutserumtherapie’’). When his data were published in 1890 [1], very little was known about the factors or mechanisms involved in immune defense. Despite this, his smart conclusion was that a human body needs some defense mechanism to fight foreign toxic substances and that these substances should be present in the blood – and therefore can be prepared from serum and used for therapy against toxins or infections. His idea worked, and the success allowed him to found the first ‘‘biotech’’ company devoted to antibody-based therapy in 1904 – using his Nobel Prize money as ‘‘venture capital.’’ The company is still active in the business today as part of CSLBehring. In 1908, Paul Ehrlich, the father of hematology [2] and the first consistent concept of immunology (‘‘lateral chain theory,’’ Figure 1.1d [3]), was awarded the second Nobel Prize related to antibody therapeutics for his groundbreaking work on serum, ‘‘particularly to the valency determination of sera preparations.’’ Ehrlich laid the foundations of antibody generation by performing systematic research on immunization schedules and their efficiency, and he was the first to describe different immunoglobulin subclasses. He also coined the phrases ‘‘passive vaccination’’ and ‘‘active vaccination.’’ His lateral chain theory (‘‘Seitenketten,’’ sometimes misleadingly translated to ‘‘side-chain theory’’) postulated chemical receptors produced by blood cells that fitted intruding toxins (antigens). -

Journal Pre-Proof
Journal Pre-proof Neutralizing monoclonal antibodies for COVID-19 treatment and prevention Juan P. Jaworski PII: S2319-4170(20)30209-2 DOI: https://doi.org/10.1016/j.bj.2020.11.011 Reference: BJ 374 To appear in: Biomedical Journal Received Date: 2 September 2020 Revised Date: 6 November 2020 Accepted Date: 22 November 2020 Please cite this article as: Jaworski JP, Neutralizing monoclonal antibodies for COVID-19 treatment and prevention, Biomedical Journal, https://doi.org/10.1016/j.bj.2020.11.011. This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. © 2020 Chang Gung University. Publishing services by Elsevier B.V. TITLE: Neutralizing monoclonal antibodies for COVID-19 treatment and prevention Juan P. JAWORSKI Consejo Nacional de Investigaciones Científicas y Técnicas, Buenos Aires, Argentina Instituto Nacional de Tecnología Agropecuaria, Buenos Aires, Argentina KEYWORDS: SARS-CoV-2, Coronavirus, Monoclonal Antibody, mAb, Prophylaxis, Treatment CORRESPONDING AUTHOR: Dr. Juan Pablo Jaworski, DVM, MSc, PhD. Consejo Nacional de Investigaciones Científicas y Técnicas Instituto de Virología, Instituto Nacional de Tecnología Agropecuaria Las Cabañas y de los Reseros (S/N), Hurlingham (1686), Buenos Aires, Argentina Tel / Fax: 054-11-4621-1447 (int:3400) [email protected] ABSTRACT The SARS-CoV-2 pandemic has caused unprecedented global health and economic crises. -

The Anti-Pseudomonas Aeruginosa Antibody Panobacumab Is Efficacious on Acute Pneumonia in Neutropenic Mice and Has Additive Effects with Meropenem
The Anti-Pseudomonas aeruginosa Antibody Panobacumab Is Efficacious on Acute Pneumonia in Neutropenic Mice and Has Additive Effects with Meropenem Thomas Secher1,3*¤a☯, Stefanie Fas2¤b☯, Louis Fauconnier1,3¤c☯, Marieke Mathieu1,3, Oliver Rutschi2, Bernhard Ryffel1,3, Michael Rudolf2 1 Université d’Orléans and Centre National de la Recherche Scientifique, Unité Mixte de Recherche, Orléans, France, 2 Kenta Biotech AG, Schlieren, Switzerland, 3 Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, Republic of South Africa Abstract Pseudomonas aeruginosa (P. aeruginosa) infections are associated with considerable morbidity and mortality in immunocompromised patients due to antibiotic resistance. Therefore, we investigated the efficacy of the anti-P. aeruginosa serotype O11 lipopolysaccharide monoclonal antibody Panobacumab in a clinically relevant murine model of neutropenia induced by cyclophosphamide and in combination with meropenem in susceptible and meropenem resistant P. aeruginosa induced pneumonia. We observed that P. aeruginosa induced pneumonia was dramatically increased in neutropenic mice compared to immunocompetent mice. First, Panobacumab significantly reduced lung inflammation and enhanced bacterial clearance from the lung of neutropenic host. Secondly, combination of Panobacumab and meropenem had an additive effect. Third, Panobacumab retained activity on a meropenem resistant P. aeruginosa strain. In conclusion, the present data established that Panobacumab contributes to the clearance of P. aeruginosa in neutropenic hosts as well as in combination with antibiotics in immunocompetent hosts. This suggests beneficial effects of co-treatment even in immunocompromised individuals, suffering most of the morbidity and mortality of P. aeruginosa infections. Citation: Secher T, Fas S, Fauconnier L, Mathieu M, Rutschi O, et al. -

Pharmacokinetic-Pharmacodynamic Modelling of Systemic IL13 Blockade by Monoclonal Antibody Therapy: a Free Assay Disguised As Total
pharmaceutics Article Pharmacokinetic-Pharmacodynamic Modelling of Systemic IL13 Blockade by Monoclonal Antibody Therapy: A Free Assay Disguised as Total John Hood 1,*, Ignacio González-García 1 , Nicholas White 1, Leeron Marshall 1,2, Vincent F. S. Dubois 1 , Paolo Vicini 1,3 and Paul G. Baverel 1,4 1 Clinical Pharmacology and Quantitative Pharmacology, AstraZeneca, Cambridge CB21 6GH, UK; [email protected] (I.G.-G.); [email protected] (N.W.); [email protected] (L.M.); [email protected] (V.F.S.D.); [email protected] (P.V.); [email protected] (P.G.B.) 2 Salford Royal Foundation Trust, Salford M6 8HD, UK 3 Confo Therapeutics, 9052 Ghent, Zwijnaarde, Belgium 4 Roche Pharma Research and Early Development, Clinical Pharmacology, Pharmaceutical Sciences, Roche Innovation Center Basel F. Hoffmann-La Roche Ltd., CH-4070 Basel, Switzerland * Correspondence: [email protected]; Tel.: +44-1223-749-6288 Abstract: A sequential pharmacokinetic (PK) and pharmacodynamic (PD) model was built with Nonlinear Mixed Effects Modelling based on data from a first-in-human trial of a novel biologic, MEDI7836. MEDI7836 is a human immunoglobulin G1 lambda (IgG1λ-YTE) monoclonal antibody, Citation: Hood, J.; González-García, with an Fc modification to reduce metabolic clearance. MEDI7836 specifically binds to, and function- I.; White, N.; Marshall, L.; Dubois, ally neutralizes interleukin-13. Thirty-two healthy male adults were enrolled into a dose-escalation V.F.S.; Vicini, P.; Baverel, P.G. clinical trial. Four active doses were tested (30, 105, 300, and 600 mg) with 6 volunteers enrolled Pharmacokinetic-Pharmacodynamic per cohort. Eight volunteers received placebo as control. -

Monoclonal Antibodies As Tools to Combat Fungal Infections
Journal of Fungi Review Monoclonal Antibodies as Tools to Combat Fungal Infections Sebastian Ulrich and Frank Ebel * Institute for Infectious Diseases and Zoonoses, Faculty of Veterinary Medicine, Ludwig-Maximilians-University, D-80539 Munich, Germany; [email protected] * Correspondence: [email protected] Received: 26 November 2019; Accepted: 31 January 2020; Published: 4 February 2020 Abstract: Antibodies represent an important element in the adaptive immune response and a major tool to eliminate microbial pathogens. For many bacterial and viral infections, efficient vaccines exist, but not for fungal pathogens. For a long time, antibodies have been assumed to be of minor importance for a successful clearance of fungal infections; however this perception has been challenged by a large number of studies over the last three decades. In this review, we focus on the potential therapeutic and prophylactic use of monoclonal antibodies. Since systemic mycoses normally occur in severely immunocompromised patients, a passive immunization using monoclonal antibodies is a promising approach to directly attack the fungal pathogen and/or to activate and strengthen the residual antifungal immune response in these patients. Keywords: monoclonal antibodies; invasive fungal infections; therapy; prophylaxis; opsonization 1. Introduction Fungal pathogens represent a major threat for immunocompromised individuals [1]. Mortality rates associated with deep mycoses are generally high, reflecting shortcomings in diagnostics as well as limited and often insufficient treatment options. Apart from the development of novel antifungal agents, it is a promising approach to activate antimicrobial mechanisms employed by the immune system to eliminate microbial intruders. Antibodies represent a major tool to mark and combat microbes. Moreover, monoclonal antibodies (mAbs) are highly specific reagents that opened new avenues for the treatment of cancer and other diseases.