The Evolution of Color Vision in Nocturnal Mammals
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ctz78-02 (02) Lee Et Al.Indd 51 14 08 2009 13:12 52 Lee Et Al
Contributions to Zoology, 78 (2) 51-64 (2009) Variation in the nocturnal foraging distribution of and resource use by endangered Ryukyu flying foxes(Pteropus dasymallus) on Iriomotejima Island, Japan Ya-Fu Lee1, 4, Tokushiro Takaso2, 5, Tzen-Yuh Chiang1, 6, Yen-Min Kuo1, 7, Nozomi Nakanishi2, 8, Hsy-Yu Tzeng3, 9, Keiko Yasuda2 1 Department of Life Sciences and Institute of Biodiversity, National Cheng Kung University, Tainan 701, Taiwan 2 The Iriomote Project, Research Institute for Humanity and Nature, 671 Iriomote, Takatomi-cho, Okinawa 907- 1542, Japan 3 Hengchun Research Center, Taiwan Forestry Research Institute, Pingtung 946, Taiwan 4 E-mail: [email protected] 5 E-mail: [email protected] 6 E-mail: [email protected] 7 E-mail: [email protected] 8 E-mail: [email protected] 9 E-mail: [email protected] Key words: abundance, bats, Chiroptera, diet, figs, frugivores, habitat Abstract Contents The nocturnal distribution and resource use by Ryukyu flying foxes Introduction ........................................................................................ 51 was studied along 28 transects, covering five types of habitats, on Material and methods ........................................................................ 53 Iriomote Island, Japan, from early June to late September, 2005. Study sites ..................................................................................... 53 Bats were mostly encountered solitarily (66.8%) or in pairs (16.8%), Bat and habitat census ................................................................ -

G- and C-Banding Chromosomal Studies of Bats of the Family Emballonuridae
G- AND C-BANDING CHROMOSOMAL STUDIES OF BATS OF THE FAMILY EMBALLONURIDAE CRAIG S. HOOD AND ROBERT J. BAKER Department of Biological Sciences and The Museum, Texas Tech University, Lubbock, TX 79409 Present address of CSH: Department of Biological Sciences, Loyola University, New Orleans, LA 70118 ABSTRACT.—Extent and nature of chromosomal change among nine species representing six genera (Saccopteryx, Rhynchonycteris, Diclidurus, Balantiopteryx, Cormura, and Taphozous) were examined using data from G- and C-banded chromosomes. Heterochromatin was restricted to centromeric regions in most taxa; extensive additions of C-positive material occurred in Bal- antiopteryx and Cormura. Comparisons of G-bands of euchromatic arms revealed considerable variation in G-band pattern and imply extensive chromosomal evolution among emballonurid species. Outgroup comparisons of G-band karyotypes proposed as primitive for several families of bats failed to reveal conserved G-band patterns, thus limiting the usefulness of differentially stained chromosomal data for resolving phylogenetic relationships of the Emballonuridae. The karyotype of Cormura brevirostris includes an extraordinarily large X chromosome that is mostly euchromatic. Evolution of the X chromosome and the nature of the sex-determining system in Cormura are unclear, but the species appears to possess a unique sex chromosome mechanism. The bat family Emballonuridae contains 13 genera and about 50 species and has a broad, pantropical distribution (Corbet and Hill, 1980; Hill and Smith, 1983). From both a systematic and cytogenetic standpoint these bats represent one of the least understood chiropteran families. With the exception of a G-band figure presented in Baker et al. (1982) for Saccopteryx canes- cens, published chromosomal data are limited to nondifferentially-stained karyotypes for 13 species representing 9 genera (Baker et al., 1982, and references therein; Harada et al., 1982; Ray-Chaudhuri et al., 1971). -

Checklist of the Mammals of Indonesia
CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation i ii CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation By Ibnu Maryanto Maharadatunkamsi Anang Setiawan Achmadi Sigit Wiantoro Eko Sulistyadi Masaaki Yoneda Agustinus Suyanto Jito Sugardjito RESEARCH CENTER FOR BIOLOGY INDONESIAN INSTITUTE OF SCIENCES (LIPI) iii © 2019 RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI) Cataloging in Publication Data. CHECKLIST OF THE MAMMALS OF INDONESIA: Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation/ Ibnu Maryanto, Maharadatunkamsi, Anang Setiawan Achmadi, Sigit Wiantoro, Eko Sulistyadi, Masaaki Yoneda, Agustinus Suyanto, & Jito Sugardjito. ix+ 66 pp; 21 x 29,7 cm ISBN: 978-979-579-108-9 1. Checklist of mammals 2. Indonesia Cover Desain : Eko Harsono Photo : I. Maryanto Third Edition : December 2019 Published by: RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI). Jl Raya Jakarta-Bogor, Km 46, Cibinong, Bogor, Jawa Barat 16911 Telp: 021-87907604/87907636; Fax: 021-87907612 Email: [email protected] . iv PREFACE TO THIRD EDITION This book is a third edition of checklist of the Mammals of Indonesia. The new edition provides remarkable information in several ways compare to the first and second editions, the remarks column contain the abbreviation of the specific island distributions, synonym and specific location. Thus, in this edition we are also corrected the distribution of some species including some new additional species in accordance with the discovery of new species in Indonesia. -

The Influence of Invasive Alien Plant Control on the Foraging Habitat Quality of the Mauritian Flying Fox (Pteropus Niger)
Master’s Thesis 2017 60 ECTS Department of Ecology and Natural Resource Management The influence of invasive alien plant control on the foraging habitat quality of the Mauritian flying fox (Pteropus niger) Gabriella Krivek Tropical Ecology and Natural Resource Management ACKNOWLEDMENTS First, I would like to express my gratitude to my supervisors Torbjørn Haugaasen from INA and Vincent Florens from the University of Mauritius, who made this project possible and helped me through my whole study and during the write-up. I also want to thank to NMBU and INA for the financial support for my field work in Mauritius. I am super thankful to Maák Isti for helping with the statistical issues and beside the useful ideas and critical comments, sending me positive thoughts on hopeless days and making me believe in myself. Special thanks to Cláudia Baider from The Mauritius Herbarium, who always gave me useful advice on my drafts and also helped me in my everyday life issues. I would like to thank Prishnee for field assistance and her genuine support in my personal life as well. I am also grateful to Mr. Owen Griffiths for his permission to conduct my study on his property and to Ravi, who generously provided me transportation to the field station, whenever I needed. Super big thanks to Rikard for printing out and submitting my thesis. I am also thankful to my family for their support during my studies, fieldwork and writing, and also for supporting the beginning of a new chapter in Mauritius. Finally, a special mention goes to my love, who has been motivating me every single day not to give up on my dreams. -

A Recent Bat Survey Reveals Bukit Barisan Selatan Landscape As A
A Recent Bat Survey Reveals Bukit Barisan Selatan Landscape as a Chiropteran Diversity Hotspot in Sumatra Author(s): Joe Chun-Chia Huang, Elly Lestari Jazdzyk, Meyner Nusalawo, Ibnu Maryanto, Maharadatunkamsi, Sigit Wiantoro, and Tigga Kingston Source: Acta Chiropterologica, 16(2):413-449. Published By: Museum and Institute of Zoology, Polish Academy of Sciences DOI: http://dx.doi.org/10.3161/150811014X687369 URL: http://www.bioone.org/doi/full/10.3161/150811014X687369 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Acta Chiropterologica, 16(2): 413–449, 2014 PL ISSN 1508-1109 © Museum and Institute of Zoology PAS doi: 10.3161/150811014X687369 A recent -

Chiroptera: Pteropodidae)
Chapter 6 Phylogenetic Relationships of Harpyionycterine Megabats (Chiroptera: Pteropodidae) NORBERTO P. GIANNINI1,2, FRANCISCA CUNHA ALMEIDA1,3, AND NANCY B. SIMMONS1 ABSTRACT After almost 70 years of stability following publication of Andersen’s (1912) monograph on the group, the systematics of megachiropteran bats (Chiroptera: Pteropodidae) was thrown into flux with the advent of molecular phylogenetics in the 1980s—a state where it has remained ever since. One particularly problematic group has been the Austromalayan Harpyionycterinae, currently thought to include Dobsonia and Harpyionycteris, and probably also Aproteles.Inthis contribution we revisit the systematics of harpyionycterines. We examine historical hypotheses of relationships including the suggestion by O. Thomas (1896) that the rousettine Boneia bidens may be related to Harpyionycteris, and report the results of a series of phylogenetic analyses based on new as well as previously published sequence data from the genes RAG1, RAG2, vWF, c-mos, cytb, 12S, tVal, 16S,andND2. Despite a striking lack of morphological synapomorphies, results of our combined analyses indicate that Boneia groups with Aproteles, Dobsonia, and Harpyionycteris in a well-supported, expanded Harpyionycterinae. While monophyly of this group is well supported, topological changes within this clade across analyses of different data partitions indicate conflicting phylogenetic signals in the mitochondrial partition. The position of the harpyionycterine clade within the megachiropteran tree remains somewhat uncertain. Nevertheless, biogeographic patterns (vicariance-dispersal events) within Harpyionycterinae appear clear and can be directly linked to major biogeographic boundaries of the Austromalayan region. The new phylogeny of Harpionycterinae also provides a new framework for interpreting aspects of dental evolution in pteropodids (e.g., reduction in the incisor dentition) and allows prediction of roosting habits for Harpyionycteris, whose habits are unknown. -
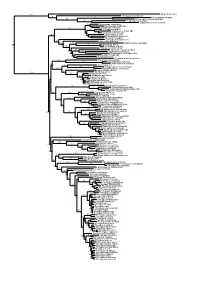
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Ecological Assessments in the B+WISER Sites
Ecological Assessments in the B+WISER Sites (Northern Sierra Madre Natural Park, Upper Marikina-Kaliwa Forest Reserve, Bago River Watershed and Forest Reserve, Naujan Lake National Park and Subwatersheds, Mt. Kitanglad Range Natural Park and Mt. Apo Natural Park) Philippines Biodiversity & Watersheds Improved for Stronger Economy & Ecosystem Resilience (B+WISER) 23 March 2015 This publication was produced for review by the United States Agency for International Development. It was prepared by Chemonics International Inc. The Biodiversity and Watersheds Improved for Stronger Economy and Ecosystem Resilience Program is funded by the USAID, Contract No. AID-492-C-13-00002 and implemented by Chemonics International in association with: Fauna and Flora International (FFI) Haribon Foundation World Agroforestry Center (ICRAF) The author’s views expressed in this publication do not necessarily reflect the views of the United States Agency for International Development or the United States Government. Ecological Assessments in the B+WISER Sites Philippines Biodiversity and Watersheds Improved for Stronger Economy and Ecosystem Resilience (B+WISER) Program Implemented with: Department of Environment and Natural Resources Other National Government Agencies Local Government Units and Agencies Supported by: United States Agency for International Development Contract No.: AID-492-C-13-00002 Managed by: Chemonics International Inc. in partnership with Fauna and Flora International (FFI) Haribon Foundation World Agroforestry Center (ICRAF) 23 March -

The Case of the Endangered Ryukyu Flying Fox
Public awareness and perceptual factors in the conservation of Title elusive species: The case of the endangered Ryukyu flying fox Vincenot, Christian Ernest; Collazo, Anja Maria; Wallmo, Author(s) Kristy; Koyama, Lina Citation Global Ecology and Conservation (2015), 3: 526-540 Issue Date 2015-01 URL http://hdl.handle.net/2433/196069 © 2015 The Authors. Published by Elsevier B.V. This is an Right open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/). Type Journal Article Textversion publisher Kyoto University Global Ecology and Conservation 3 (2015) 526–540 Contents lists available at ScienceDirect Global Ecology and Conservation journal homepage: www.elsevier.com/locate/gecco Original research article Public awareness and perceptual factors in the conservation of elusive species: The case of the endangered Ryukyu flying fox Christian Ernest Vincenot a,∗, Anja Maria Collazo b, Kristy Wallmo c, Lina Koyama a a Department of Social Informatics, Graduate School of Informatics, Kyoto University, Kyoto, Japan b Graduate School of Human and Environmental Studies, Kyoto University, Kyoto, Japan c National Marine Fisheries Service, National Oceanographic and Atmospheric Administration, Silver Spring, MD, USA article info a b s t r a c t Article history: The success of biological conservation initiatives is not solely reliant on the collection of Received 23 October 2014 ecological information, but equally on public adherence to protection programs. Awareness Received in revised form 7 February 2015 and perception of target species condition the intensity and orientation of public involve- Accepted 7 February 2015 ment in conservation initiatives. Their evaluation is critical in the case of elusive animals, Available online 13 February 2015 for which incertitude surrounding public attitude is maximized. -

A Parasite of Balantiopteryx Plicata (Chiroptera) in Mexico
Parasite 2013, 20,47 Ó J.M. Caspeta-Mandujano et al., published by EDP Sciences, 2013 DOI: 10.1051/parasite/2013047 urn:lsid:zoobank.org:pub:E2016916-65BF-4F47-A68B-2F0E16B6710A Available online at: www.parasite-journal.org RESEARCH ARTICLE OPEN ACCESS Pterygodermatites (Pterygodermatites) mexicana n. sp. (Nematoda: Rictulariidae), a parasite of Balantiopteryx plicata (Chiroptera) in Mexico Juan Manuel Caspeta-Mandujano1,2,*, Francisco Agustı´n Jime´nez3, Jorge Luis Peralta-Rodrı´guez4, and Jose´ Antonio Guerrero5 1 Laboratorio de Parasitologı´a de Animales Silvestres, Facultad de Ciencias Biolo´gicas. Av. Universidad No. 1001, Col. Chamilpa, C.P. 62210, Cuernavaca, Morelos, Me´xico 2 Centro de Investigaciones Biolo´gicas, Universidad Auto´noma del Estado de Morelos, Av. Universidad No. 1001, Col. Chamilpa, C.P. 62210, Cuernavaca, Morelos, Me´xico 3 Department of Zoology, Southern Illinois University, Carbondale, Illinois 62901-6501, USA 4 Facultad de Ciencias Agropecuarias, Universidad Auto´noma del Estado de Morelos, Av. Universidad No. 1001, Col. Chamilpa, C.P. 62210, Cuernavaca, Morelos, Me´xico 5 Laboratorio de Sistema´tica y Morfologı´a, Facultad de Ciencias Biolo´gicas, Universidad Auto´noma del Estado de Morelos. Av. Universidad No. 1001, Col. Chamilpa, C.P. 62210, Cuernavaca, Morelos, Me´xico Received 18 July 2013, Accepted 10 November 2013, Published online 26 November 2013 Abstract – A new species of nematode, Pterygodermatites (Pterygodermatites) mexicana n. sp., is described based on specimens recovered from the intestine of the gray sac-winged bat, Balantiopteryx plicata (Chiroptera, Emballonuri- dae), from the Biosphere Reserve ‘‘Sierra de Huautla’’ in the state of Morelos, Mexico. This is the second species in the genus described from bats in the New World, since most of the rictaluriids reported in these hosts belong to the closely related genus Rictularia Froelich, 1802. -

Bat Count 2003
BAT COUNT 2003 Working to promote the long term, sustainable conservation of globally threatened flying foxes in the Philippines, by developing baseline population information, increasing public awareness, and training students and protected area managers in field monitoring techniques. 1 A Terminal Report Submitted by Tammy Mildenstein1, Apolinario B. Cariño2, and Samuel Stier1 1Fish and Wildlife Biology, University of Montana, USA 2Silliman University and Mt. Talinis – Twin Lakes Federation of People’s Organizations, Diputado Extension, Sibulan, Negros Oriental, Philippines Photo by: Juan Pablo Moreiras 2 EXECUTIVE SUMMARY Large flying foxes in insular Southeast Asia are the most threatened of the Old World fruit bats due to deforestation, unregulated hunting, and little conservation commitment from local governments. Despite the fact they are globally endangered and play essential ecological roles in forest regeneration as seed dispersers and pollinators, there have been only a few studies on these bats that provide information useful to their conservation management. Our project aims to promote the conservation of large flying foxes in the Philippines by providing protected area managers with the training and the baseline information necessary to design and implement a long-term management plan for flying foxes. We focused our efforts on the globally endangered Philippine endemics, Acerodon jubatus and Acerodon leucotis, and the bats that commonly roost with them, Pteropus hypomelanus, P. vampyrus lanensis, and P. pumilus which are thought to be declining in the Philippines. Local participation is an integral part of our project. We conducted the first national training workshop on flying fox population counts and conservation at the Subic Bay area. -

INSIGHTS INTO RELATIONSHIPS AMONG RODENT LINEAGES BASED on MITOCHONDRIAL GENOME SEQUENCE DATA a Dissertation by LAURENCE JOHN FR
INSIGHTS INTO RELATIONSHIPS AMONG RODENT LINEAGES BASED ON MITOCHONDRIAL GENOME SEQUENCE DATA A Dissertation by LAURENCE JOHN FRABOTTA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY December 2005 Major Subject: Zoology INSIGHTS INTO RELATIONSHIPS AMONG RODENT LINEAGES BASED ON MITOCHONDRIAL GENOME SEQUENCE DATA A Dissertation by LAURENCE JOHN FRABOTTA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Approved by: Chair of Committee, Rodney L. Honeycutt Committee Members, James B. Woolley John W. Bickham James R. Manhart Head of Department, Vincent M. Cassone December 2005 Major Subject: Zoology iii ABSTRACT Insights into Relationships among Rodent Lineages Based on Mitochondrial Genome Sequence Data. (December 2005) Laurence John Frabotta, B.S.; M.S., California State University, Long Beach Chair of Advisory Committee: Dr. Rodney L. Honeycutt This dissertation has two major sections. In Chapter II, complete mitochondrial (mt DNA) genome sequences were used to construct a hypothesis for affinities of most major lineages of rodents that arose quickly in the Eocene and were well established by the end of the Oligocene. Determining the relationships among extant members of such old lineages can be difficult. Two traditional schemes on subordinal classification of rodents have persisted for over a century, dividing rodents into either two or three suborders, with relationships among families or superfamilies remaining problematic. The mtDNA sequences for four new rodent taxa (Aplodontia, Cratogeomys, Erethizon, and Hystrix), along with previously published Euarchontoglires taxa, were analyzed under parsimony, likelihood, and Bayesian criteria.