The African Turquoise Killifish: a Model for Exploring Vertebrate Aging and Diseases in the Fast Lane
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deterministic Shifts in Molecular Evolution Correlate with Convergence to Annualism in Killifishes
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.09.455723; this version posted August 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Deterministic shifts in molecular evolution correlate with convergence to annualism in killifishes Andrew W. Thompson1,2, Amanda C. Black3, Yu Huang4,5,6 Qiong Shi4,5 Andrew I. Furness7, Ingo, Braasch1,2, Federico G. Hoffmann3, and Guillermo Ortí6 1Department of Integrative Biology, Michigan State University, East Lansing, Michigan 48823, USA. 2Ecology, Evolution & Behavior Program, Michigan State University, East Lansing, MI, USA. 3Department of Biochemistry, Molecular Biology, Entomology, & Plant Pathology, Mississippi State University, Starkville, MS 39759, USA. 4Shenzhen Key Lab of Marine Genomics, Guangdong Provincial Key Lab of Molecular Breeding in Marine Economic Animals, BGI Marine, Shenzhen 518083, China. 5BGI Education Center, University of Chinese Academy of Sciences, Shenzhen 518083, China. 6Department of Biological Sciences, The George Washington University, Washington, DC 20052, USA. 7Department of Biological and Marine Sciences, University of Hull, UK. Corresponding author: Andrew W. Thompson, [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.08.09.455723; this version posted August 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: The repeated evolution of novel life histories correlating with ecological variables offer opportunities to test scenarios of convergence and determinism in genetic, developmental, and metabolic features. Here we leverage the diversity of aplocheiloid killifishes, a clade of teleost fishes that contains over 750 species on three continents. -

The Mystery of the Banded Killifish Fundulus Diaphanus Population Explosion: Where Did They All Come From?
The Mystery of the Banded KillifishFundulus ( diaphanus) Population Explosion: Where Did They All Come from? Philip W. Willink, Jeremy S. Tiemann, Joshua L. Sherwood, Eric R. Larson, Abe Otten, Brian Zimmerman 3 American Currents Vol. 44, No. 4 THE MYSTERY OF THE BANDED KILLIFISH FUNDULUS DIAPHANUS POPULATION EXPLOSION: WHERE DID THEY ALL COME FROM? Philip W. Willink, Jeremy S. Tiemann, Joshua L. Sherwood, La Grange Park, IL Illinois Natural History Survey Illinois Natural History Survey Eric R. Larson, Abe Otten, Brian Zimmerman University of Illinois at Scott Community The Ohio State University Urbana-Champaign College Stream and River Ecology Lab Banded Killifish Fundulus diaphanus are no strangers to NAN- and have not been seen since, they were only known from a hand- FAers. Over the past several years, there have been multiple arti- ful of inland lakes in the far northeastern corner the state (Fig. 1). cles in American Currents covering their distribution (Hatch 2015; Even there, population numbers were low. Schmidt 2016a, 2018; Olson and Schmidt 2018; Li 2019), stocking So it was with great excitement that in the early 2000s Illinois to restore populations (Bland 2013; Schmidt 2014), and appear- ichthyologists started to find more and more presumed Western ance in a hatchery (Schmidt 2016b). Their range extends from the Banded Killifish in Lake Michigan (Willink et al. 2018). They Canadian Maritime provinces south along the Atlantic coast to were even showing up in downtown Chicago (Willink 2011). It the Carolinas, as well as westward through the Great Lakes region was hoped that this range expansion was evidence of an uncom- to the upper Mississippi watershed. -

Gene Expression After Freshwater Transfer in Gills and Opercular Epithelia of Killifish: Insight Into Divergent Mechanisms of Ion Transport Graham R
The Journal of Experimental Biology 208, 2719-2729 2719 Published by The Company of Biologists 2005 doi:10.1242/jeb.01688 Gene expression after freshwater transfer in gills and opercular epithelia of killifish: insight into divergent mechanisms of ion transport Graham R. Scott1,*, James B. Claiborne2, Susan L. Edwards2,3, Patricia M. Schulte1 and Chris M. Wood4 1Department of Zoology, University of British Columbia, Vancouver BC, Canada V6T 1Z4, 2Department of Biology, Georgia Southern University, Statesboro, GA 30460-8042, USA, 3Department of Physiology and Pharmacology, James Cook University, Cairns, QLD 4879, Australia and 4Department of Biology, McMaster University, Hamilton ON, Canada L8S 4K1 *Author for correspondence (e-mail: [email protected]) Accepted 17 May 2005 Summary We have explored the molecular basis for differences in epithelium, NHE2 was not expressed; furthermore, physiological function between the gills and opercular Na+,K+-ATPase activity was unchanged after transfer to epithelium of the euryhaline killifish Fundulus freshwater, CA2 mRNA levels decreased, and NHE3 levels heteroclitus. These tissues are functionally similar in increased. Consistent with their functional similarities in seawater, but in freshwater the gills actively absorb Na+ seawater, killifish gills and opercular epithelium expressed – + + + + – but not Cl , whereas the opercular epithelium actively Na ,K -ATPase α1a, Na ,K ,2Cl cotransporter 1 absorbs Cl– but not Na+. These differences in freshwater (NKCC1), cystic fibrosis transmembrane conductance physiology are likely due to differences in absolute levels regulator (CFTR) Cl– channel and the signalling protein of gene expression (measured using real-time PCR), as 14-3-3a at similar absolute levels. Furthermore, NKCC1 several proteins important for Na+ transport, namely and CFTR were suppressed equally in each tissue after Na+,H+-exchanger 2 (NHE2), carbonic anhydrase 2 (CA2), freshwater transfer, and 14-3-3a mRNA increased in both. -
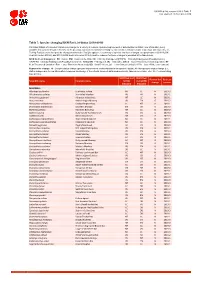
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Draft Risk Assessment Report Nothobranchius Furzeri (Turquoise
APPLICATION TO AMEND THE LIST OF SPECIMENS SUITABLE FOR LIVE IMPORT Draft Risk Assessment Report 1. Taxonomy of the species a. Family name: Aplocheilidae b. Genus name: Nothobranchius c. Species: N. furzeri d. Subspecies: No e. Taxonomic Reference: http://www.uniprot.org/taxonomy/105023 f. Common Names: Turquoise killifish g. Is the species a genetically-modified organism (GMO)? No 2. Status of the species under CITES Species is not listed in the CITES Appendices. 3. Ecology of the species a. Longevity: what is the average lifespan of the species in the wild and in captivity? The average lifespan of the species in captivity is 13 weeks (1). Wild derived N. furzeri from three different habitats showed a maximum lifespan of 25-32 weeks (1). b. What is the maximum length and weight that the species attains? Provide information on the size and weight range for males and females of the species. As per the original formal description of the species, the standard length of N. furzeri from a wild population ranged from 2.5cm to 4.4cm (2). The maximum length the species can attain is 7cm (3). The mean maximal body weight reported for males is 3.8 ± 1.1 g (range: 0.5 – 6.7 g) and 2.2 ± 0.6 g (range: 0.2 - 3.6 g) for females (4). Growth rates of Nothobranchius in the wild have been reported to be similar to those in captivity (5). c. Discuss the identification of the individuals in this species, including if the sexes of the species are readily distinguishable, and if the species is difficult to distinguish from other species. -

Gill Trematodes (Flukes) in Wild-Caught Killifish (<I>Fundulus
NOTES Gill Trematodes (Flukes) in Wild-Caught Killifish (Fundulus heteroclitus) DAVID R. GOULDING, BS, RLAT,1* TERRY L. BLANKENSHIP-PARIS, DVM, MS, DIPLOMATE, ACLAM,1 GREGORY A. LEWBART, MS, VMD, DIPLOMATE, ACZM,2 PAGE H. MYERS, BS, LATG,1 TRACY K. DEMIANENKO, BS, RLAT,1 JAMES A. CLARK, LATG,1 AND DIANE B. FORSYTHE, DVM, DIPLOMATE, ACLAM1 Three wild caught killifish (Fundulus heteroclitus) on an Institutional Animal Care and Use Committee-approved protocol were found dead within 2 days after being received. The fish were housed in two separate aquaria. Aquarium water was evaluated, and pH, salinity, ammonia, nitrate, and nitrite levels were within acceptable parameters. Several remaining fish appeared to be slow-moving and were presented for necropsy. Multiple, scattered, ulcerated skin lesions (diameter, 1 to 5 mm) were noted at necropsy and were cultured. No pathogenic bacteria were isolated. Wet-mount samples of the gills revealed multiple cysts at the gill margins, each containing a motile organism. No other gill parasites were detected. A diagnosis of trematodiasis was made. The cysts were identified as encysted metacercariae of a digenetic trematode. We surmise that the large numbers of gill flukes combined with the stress of recent shipment likely caused the observed morbidity and mortality. Sixty-two wild-caught killifish (Fundulus heteroclitus) obtained from Duke Marine Center (Beaufort, N.C.) were group-housed upon receipt in two 38-gallon aquaria containing conditioned brack- ish water. Thirty-six fish were placed in tank A, and 26 fish were placed in tank B. One day after arrival, two fish were found dead in tank B, and 2 days after arrival, one fish was found dead in tank A. -

Compare and Contrast the Water Environment Between Death Valley Pupfish Specie and Devil’S Hole Pupfish Specie
Compare and Contrast the Water environment between Death Valley Pupfish Specie and Devil’s Hole Pupfish Specie By Roy Tianran Gao 1 Table of Contents Title page 1 Abstract 3 Introduction and Background 3 Water Temperature 4 Salinity 6 Water Level 7 Conservation 10 Conclusion 11 References 12 2 ABSTRACT The two types of pupfish (Cyprinodon) in Death Valley National Park are Death Valley pupfish and Devil’s Hole pupfish. Death Valley pupfish has been existed over 10,000 years and Devil’s Hole pupfish has been existed for over 20,000 years. Both of the pupfishes are endangered species. The average number of Death Valley pupfish has decreased by about 100 since 1990s, and the number of Devil’s Hole pupfish has decreased by 400 since 1995. Comparing the water level, water temperature and the water salinity between the two species of pupfish would help to define the living requirements and reason of decreasing population. The research toward the result is based on 7 journal articles, 4 websites, and 1 book. As the result shows, Death Valley Pupfish and Devil’s Hole Pupfish live in different water environments and functioned differently. Understanding the water environment of the two types of pupfishes will help people building new habitats for pupfishes and increase their population so that would be possible to avoid the extinction of pupfishes from the earth. INTRODUCTION AND BACKGROUND Pupfish is a small killifish in the Southwest of America. There are five pupfish species in Death Valley which are Armargosa pupfish, Saratoga Pupfish, Devil’s Hole pupfish, Death Valley pupfish, and Cotton ball Marsh pupfish (National Park Service, 2008). -

Cyprinodontiformes, Nothobranchiidae)
COMPARATIVE A peer-reviewed open-access journal CompCytogen 10(3): Divergent439–445 (2016) karyotypes of the annual killifish genusNothobranchius ... 439 doi: 10.3897/CompCytogen.v10i3.9863 SHORT COMMUNICATION Cytogenetics http://compcytogen.pensoft.net International Journal of Plant & Animal Cytogenetics, Karyosystematics, and Molecular Systematics Divergent karyotypes of the annual killifish genus Nothobranchius (Cyprinodontiformes, Nothobranchiidae) Eugene Krysanov1, Tatiana Demidova1, Bela Nagy2 1 Severtsov Institute of Ecology and Evolution, Russian Academy of Sciences, Leninsky prospect, Moscow, 119071 Russia 2 30, rue du Mont Ussy, 77300 Fontainebleau, France Corresponding author: Eugene Krysanov ([email protected]) Academic editor: I. Kuznetsova | Received 13 July 2016 | Accepted 18 August 2016 | Published 16 September 2016 http://zoobank.org/516FBC12-825D-46E0-BAAA-25ABA00F8607 Citation: Krysanov E, Demidova T, Nagy B (2016) Divergent karyotypes of the annual killifish genus Nothobranchius (Cyprinodontiformes; Nothobranchiidae). Comparative Cytogenetics 10(3): 439–445. doi: 10.3897/CompCytogen. v10i3.9863 Abstract Karyotypes of two species of the African annual killifish genus Nothobranchius Peters, 1868, N. brieni Poll, 1938 and Nothobranchius sp. from Kasenga (D.R. Congo) are described. Both species displayed dip- loid chromosome number 2n = 49/50 for males and females respectively with multiple-sex chromosome system type X1X2Y/X1X1X2X2. The karyotypes of studied species are considerably different from those previously reported for the genus Nothobranchius and similar to the Actinopterygii conservative karyotype. Keywords Africa, chromosome number, karyotype, killifish, Nothobranchius Introduction Annual killifishes belonging to the genusNothobranchius Peters, 1868 are mainly dis- tributed in eastern Africa but several species are found in central Africa (Wildekamp 2004). They inhabit temporary pools that dry out during the dry season and have spe- cific adaptations for extreme environments. -

Recovery Plan Endangered and Species Nevada
RECOVERYPLAN FOR THE ENDANGEREDAND THREATENED SPECIES OF&H MEADOWS, NEVADA Prepared by Don W. Sada U.S. Fish and Wildlife Service Reno, Nevada RECOVERY PLAN FOR THE ENDANGERED AND THREATENED SPECIES OF ASH MEADOWS, NEVADA Prepared By Don W. Sada U.S. Fish and Wildlife Service Reno, Nevada for the U.S. Fish and Wildlife Service Portland, Oregon ~FP2 3 ‘:XN Date This plan covers the following federally listed species in Ash Meadows, Nevada and California: Devil’s Hole pupfish, Warm Springs pupfish, Ash Meadows Arnargosa pupfish, Ash Meadows speckled dace, Ash Meadows naucorid, Ash Meadows blazing star, Ash Meadows ivesia, Ainargosa niterwort, Spring-loving centaury, Ash Meadows sunray, Ash Meadows inilk-vetch, and Ash Meadows guxnplant. THIS IS THE COMPLETED ASH MEADOWS SPECIES RECOVERY PLAN. IT HAS BEEN APPROVED BY THE U.S. FISH AND WILDLIFE SERVICE. IT DOES NOT NECESSARILY REPRESENT OFFICIAL POSITIONS OR APPROVALS OF COOPERATING AGENCIES (AND IT DOES NOT NECESSARILY REPRESENT THE VIEWS OF ALL INDIVIDUALS) WHO PLAYED THE KEY ROLE IN PREPARING THIS PLAN. THIS PLAN IS SUBJECT TO MODIFICATION AS DICTATED BY NEW FINDINGS AND CHANGES IN SPECIES STATUS, AND COMPLETION OF TASKS DESCRIBED IN THE PLAN. GOALS AND OBJECTIVES WILL BE ATTAINED AND FUNDS EXPENDED CONTINGENT UPON APPROPRIATIONS, PRIORITIES, AND OTHER BUDGETARY CONSTRAINTS. LITERATURE CITATION SHOULD READ AS FOLLOWS U.S. Fish and Wildlife Service. 1990. Recovery plan for the endangered and threatened species of Ash Meadows, Nevada. U.S. Fish and Wildlife Service, Portland, Oregon. 123 pp. Additional copies may be obtained from Fish and Wildlife Reference Service 5430 Grosvenor Lane, Suite 110 Bethesda, Maryland 20814 Telephone: 301-492-6403 1-800-582-3421 : ACKNOWLEDGMENTS: This plan results from the efforts of many who spent considerable time and energy to prevent the destruction of Ash Meadows and the extinction of its diverse endemic biota. -
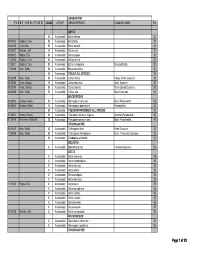
MAS BAP Species Point Table
ANABANTOID F I R S T V E R I F I E D CLASS GROUP GENUS/SPECIES COMMON NAME Pts BETTA B Anabantoid Betta edithae 10 05/01/03 Wojtech, Tom B Anabantoid Betta falax 10 09/24/95 Cram, Dan B Anabantoid Betta imbellis 10 02/06/07 Michels, Jeff B Anabantoid Betta picta 10 09/28/02 Rogne, Eric B Anabantoid Betta pugnax 10 11/25/00 Wojtech, Tom B Anabantoid Betta pulchra 10 11/25/01 Wojtech, Tom B Anabantoid Betta smaragdina Emerald Betta. 10 11/03/88 Kern, Wally B Anabantoid Betta splendens 10 B Anabantoid COLISA ALL SPECIES 10 07/23/89 Kern, Wally B Anabantoid Colisa chuna Honey Dwarf Gourami. 10 02/28/96 Tews, Woody B Anabantoid Colisa fasciatus Giant Gourami. 10 03/13/95 Tews, Woody B Anabantoid Colisa labiosa Thick-Lipped Gourami. 10 06/28/88 Kern, Wally B Anabantoid Colisa lalia Dwarf Gourami. 10 MACROPODUS 09/28/02 Korotev, Kevin B Anabantoid Macropodus concolor Black Paradisefish. 10 04/28/02 Korotev, Kevin B Anabantoid Macropodus opercularis Paradisefish. 10 B Anabantoid PSEUDOSPHROMENUS ALL SPECIES 10 01/25/03 Korotev, Kevin B Anabantoid Pseudosphromenus cupanis Spiketail Paradisefish. 10 01/28/96 Kelnhofer,, Rob & El B Anabantoid Pseudosphromenuspy dayi Day'sy Paradisefish. 10 TRICHOGASTER 05/11/87 Kern, Wally B Anabantoid Trichogaster leeri Pearl Gourami. 10 12/05/88 Kern, Wally B Anabantoid Trichogaster trichopterus Blue, Three-spot, Gourami. 10 B Anabantoid Trichopsis schalleri 10 BELONTIA C Anabantoid Belontia signata Combtail Gourami. 15 BETTA C Anabantoid Betta akarensis 15 C Anabantoid Betta anabantoides 15 C Anabantoid Betta -

Fundulus Diaphanus) Ecological Risk Screening Summary
Banded Killifish (Fundulus diaphanus) Ecological Risk Screening Summary U.S. Fish & Wildlife Service, December 2016 Revised, November 2017 Web version, 8/16/2018 Photo: hjnugent(iNaturalist). Licensed under Creative Commons BY-NC 4.0. Available: http://eol.org/data_objects/33497673. (November 1, 2017). 1 Native Range and Status in the United States Native Range From Fuller and Neilson (2016): “Atlantic Slope drainages from Newfoundland to Pee Dee River, South Carolina; St. Lawrence- Great Lakes and Mississippi River basins from Quebec to Manitoba, and south to southern Pennsylvania, northern Illinois, and northeastern Nebraska. Two subspecies with intergrades in 1 St. Lawrence and Lake Erie drainages: F[undulus]. d[iaphanus]. diaphanus on the Atlantic Slope, F. d. menona in the rest of the range (Page and Burr 1991).” From NatureServe (2013): “Range includes Atlantic Slope drainages from the Peedee River, South Carolina, north to the Maritime Provinces and Newfoundland; St. Lawrence-Great Lakes and Mississippi River basins from Quebec to Manitoba, south to southern Pennsylvania, northern Illinois, and northeastern Nebraska (Page and Burr 2011).” Status in the United States The native range of this species is within the United States (Fuller and Neilson 2016). From Fuller and Neilson (2016): “The species is established in Silvio O. Conte National Fish and Wildlife Refuge, Franklin County, Massachussetts [sic] (USFWS 2005). The eastern subspecies F. d. diaphanus was introduced into Lake Erie, the Little Miami drainage, and several areas in the upper Ohio River basin in Ohio, and into the Ohio and Beaver River drainages (Raney 1938, cited in Trautman 1981), the Monongahela drainage (Hocutt et al. -

Solar UVB-Induced DNA Damage and Photoenzymatic DNA Repair in Antarctic Zooplankton (Ozone Depletion͞dna Damage and Repair͞photolyase͞marine Ecosystems)
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 1258–1263, February 1997 Ecology Solar UVB-induced DNA damage and photoenzymatic DNA repair in antarctic zooplankton (ozone depletionyDNA damage and repairyphotolyaseymarine ecosystems) KIRK D. MALLOY*, MOLLY A. HOLMAN*, DAVID MITCHELL†, AND H. WILLIAM DETRICH III*‡ *Department of Biology, Northeastern University, Boston, MA 02115; and †Department of Carcinogenesis, The University of Texas M. D. Anderson Cancer Research Center, Smithville, TX 78957 Communicated by George Somero, Stanford University, Pacific Grove, CA, December 13, 1996 (received for review August 18, 1996) ABSTRACT The detrimental effects of elevated intensi- of marine organisms in coastal regions and the upper photic ties of mid-UV radiation (UVB), a result of stratospheric zone of open oceans may be affected deleteriously by the ozone depletion during the austral spring, on the primary projected long-term increase in UVB flux (16–20). producers of the Antarctic marine ecosystem have been well Although UVB damages most biological macromolecules, documented. Here we report that natural populations of including lipids, proteins, and nucleic acids (21, 22), the Antarctic zooplankton also sustain significant DNA damage principal cause of UV-induced mutation, degeneration, [measured as cyclobutane pyrimidine dimers (CPDs)] during andyor death in animal cells is modification of DNA (23). The periods of increased UVB flux. This is the first direct evidence most significant DNA lesions generated by UVB are cyclobu- that increased solar UVB may result in damage to marine tane pyrimidine dimers (CPDs), which constitute '70–90% of organisms other than primary producers in Antarctica. The the aberrant DNA photoproducts (23). CPDs increase linearly extent of DNA damage in pelagic icefish eggs correlated with with UVB exposure (23, 24), but the dose–response relation- daily incident UVB irradiance, reflecting the difference be- ship varies significantly between different species and taxa tween acquisition and repair of CPDs.