Cyprinodontiformes, Nothobranchiidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A New Genus of Miniature Cynolebiasine from the Atlantic
64 (1): 23 – 33 © Senckenberg Gesellschaft für Naturforschung, 2014. 16.5.2014 A new genus of miniature cynolebiasine from the Atlantic Forest and alternative biogeographical explanations for seasonal killifish distribution patterns in South America (Cyprinodontiformes: Rivulidae) Wilson J. E. M. Costa Laboratório de Sistemática e Evolução de Peixes Teleósteos, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Caixa Postal 68049, CEP 21944 – 970, Rio de Janeiro, Brasil; wcosta(at)acd.ufrj.br Accepted 21.ii.2014. Published online at www.senckenberg.de/vertebrate-zoology on 30.iv.2014. Abstract The analysis of 78 morphological characters for 16 species representing all the lineages of the tribe Cynopoecilini and three out-groups, indicates that the incertae sedis miniature species ‘Leptolebias’ leitaoi Cruz & Peixoto is the sister group of a clade comprising the genera Leptolebias, Campellolebias, and Cynopoecilus, consequently recognised as the only member of a new genus. Mucurilebias gen. nov. is diagnosed by seven autapomorphies: eye occupying great part of head side, low number of caudal-fin rays (21), distal portion of epural much broader than distal portion of parhypural, an oblique red bar through opercle in both sexes, isthmus bright red in males, a white stripe on the distal margin of the dorsal fin in males, and a red stripe on the distal margin of the anal fin in males.Mucurilebias leitaoi is an endangered seasonal species endemic to the Mucuri river basin. The biogeographical analysis of genera of the subfamily Cynolebiasinae using a dispersal-vicariance, event-based parsimony approach indicates that distribution of South American killifishes may be broadly shaped by dispersal events. -

Deterministic Shifts in Molecular Evolution Correlate with Convergence to Annualism in Killifishes
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.09.455723; this version posted August 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Deterministic shifts in molecular evolution correlate with convergence to annualism in killifishes Andrew W. Thompson1,2, Amanda C. Black3, Yu Huang4,5,6 Qiong Shi4,5 Andrew I. Furness7, Ingo, Braasch1,2, Federico G. Hoffmann3, and Guillermo Ortí6 1Department of Integrative Biology, Michigan State University, East Lansing, Michigan 48823, USA. 2Ecology, Evolution & Behavior Program, Michigan State University, East Lansing, MI, USA. 3Department of Biochemistry, Molecular Biology, Entomology, & Plant Pathology, Mississippi State University, Starkville, MS 39759, USA. 4Shenzhen Key Lab of Marine Genomics, Guangdong Provincial Key Lab of Molecular Breeding in Marine Economic Animals, BGI Marine, Shenzhen 518083, China. 5BGI Education Center, University of Chinese Academy of Sciences, Shenzhen 518083, China. 6Department of Biological Sciences, The George Washington University, Washington, DC 20052, USA. 7Department of Biological and Marine Sciences, University of Hull, UK. Corresponding author: Andrew W. Thompson, [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.08.09.455723; this version posted August 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: The repeated evolution of novel life histories correlating with ecological variables offer opportunities to test scenarios of convergence and determinism in genetic, developmental, and metabolic features. Here we leverage the diversity of aplocheiloid killifishes, a clade of teleost fishes that contains over 750 species on three continents. -

The Mystery of the Banded Killifish Fundulus Diaphanus Population Explosion: Where Did They All Come From?
The Mystery of the Banded KillifishFundulus ( diaphanus) Population Explosion: Where Did They All Come from? Philip W. Willink, Jeremy S. Tiemann, Joshua L. Sherwood, Eric R. Larson, Abe Otten, Brian Zimmerman 3 American Currents Vol. 44, No. 4 THE MYSTERY OF THE BANDED KILLIFISH FUNDULUS DIAPHANUS POPULATION EXPLOSION: WHERE DID THEY ALL COME FROM? Philip W. Willink, Jeremy S. Tiemann, Joshua L. Sherwood, La Grange Park, IL Illinois Natural History Survey Illinois Natural History Survey Eric R. Larson, Abe Otten, Brian Zimmerman University of Illinois at Scott Community The Ohio State University Urbana-Champaign College Stream and River Ecology Lab Banded Killifish Fundulus diaphanus are no strangers to NAN- and have not been seen since, they were only known from a hand- FAers. Over the past several years, there have been multiple arti- ful of inland lakes in the far northeastern corner the state (Fig. 1). cles in American Currents covering their distribution (Hatch 2015; Even there, population numbers were low. Schmidt 2016a, 2018; Olson and Schmidt 2018; Li 2019), stocking So it was with great excitement that in the early 2000s Illinois to restore populations (Bland 2013; Schmidt 2014), and appear- ichthyologists started to find more and more presumed Western ance in a hatchery (Schmidt 2016b). Their range extends from the Banded Killifish in Lake Michigan (Willink et al. 2018). They Canadian Maritime provinces south along the Atlantic coast to were even showing up in downtown Chicago (Willink 2011). It the Carolinas, as well as westward through the Great Lakes region was hoped that this range expansion was evidence of an uncom- to the upper Mississippi watershed. -

Fish, Various Invertebrates
Zambezi Basin Wetlands Volume II : Chapters 7 - 11 - Contents i Back to links page CONTENTS VOLUME II Technical Reviews Page CHAPTER 7 : FRESHWATER FISHES .............................. 393 7.1 Introduction .................................................................... 393 7.2 The origin and zoogeography of Zambezian fishes ....... 393 7.3 Ichthyological regions of the Zambezi .......................... 404 7.4 Threats to biodiversity ................................................... 416 7.5 Wetlands of special interest .......................................... 432 7.6 Conservation and future directions ............................... 440 7.7 References ..................................................................... 443 TABLE 7.2: The fishes of the Zambezi River system .............. 449 APPENDIX 7.1 : Zambezi Delta Survey .................................. 461 CHAPTER 8 : FRESHWATER MOLLUSCS ................... 487 8.1 Introduction ................................................................. 487 8.2 Literature review ......................................................... 488 8.3 The Zambezi River basin ............................................ 489 8.4 The Molluscan fauna .................................................. 491 8.5 Biogeography ............................................................... 508 8.6 Biomphalaria, Bulinis and Schistosomiasis ................ 515 8.7 Conservation ................................................................ 516 8.8 Further investigations ................................................. -

Nothobranchius As a Model for Aging Studies. a Review
Volume 5, Number 2; xxx-xx, April 2014 http://dx.doi.org/10.14336/AD Review Article Nothobranchius as a model for aging studies. A review Alejandro Lucas-Sánchez 1, *, Pedro Francisco Almaida-Pagán 2, Pilar Mendiola 1, Jorge de Costa 1 1 Department of Physiology. Faculty of Biology. University of Murcia. 30100 Murcia, Spain. 2 Institute of Aquaculture, School of Natural Sciences, University of Stirling, Stirling FK9 4LA, UK. [Received October 17, 2013; Revised December 4, 2013; Accepted December 4, 2013] ABSTRACT: In recent decades, the increase in human longevity has made it increasingly important to expand our knowledge on aging. To accomplish this, the use of animal models is essential, with the most common being mouse (phylogenetically similar to humans, and a model with a long life expectancy) and Caenorhabditis elegans (an invertebrate with a short life span, but quite removed from us in evolutionary terms). However, some sort of model is needed to bridge the differences between those mentioned above, achieving a balance between phylogenetic distance and life span. Fish of the genus Nothobranchius were suggested 10 years ago as a possible alternative for the study of the aging process. In the meantime, numerous studies have been conducted at different levels: behavioral (including the study of the rest-activity rhythm), populational, histochemical, biochemical and genetic, among others, with very positive results. This review compiles what we know about Nothobranchius to date, and examines its future prospects as a true alternative to the classic models for studies on aging. Key words: Aging, Fish, killifish, Nothobranchius Aging is a multifactorial process that involves numerous To be considered a model for aging, an animal must mechanisms that operate during the normal life cycle of have a number of features. -

Gene Expression After Freshwater Transfer in Gills and Opercular Epithelia of Killifish: Insight Into Divergent Mechanisms of Ion Transport Graham R
The Journal of Experimental Biology 208, 2719-2729 2719 Published by The Company of Biologists 2005 doi:10.1242/jeb.01688 Gene expression after freshwater transfer in gills and opercular epithelia of killifish: insight into divergent mechanisms of ion transport Graham R. Scott1,*, James B. Claiborne2, Susan L. Edwards2,3, Patricia M. Schulte1 and Chris M. Wood4 1Department of Zoology, University of British Columbia, Vancouver BC, Canada V6T 1Z4, 2Department of Biology, Georgia Southern University, Statesboro, GA 30460-8042, USA, 3Department of Physiology and Pharmacology, James Cook University, Cairns, QLD 4879, Australia and 4Department of Biology, McMaster University, Hamilton ON, Canada L8S 4K1 *Author for correspondence (e-mail: [email protected]) Accepted 17 May 2005 Summary We have explored the molecular basis for differences in epithelium, NHE2 was not expressed; furthermore, physiological function between the gills and opercular Na+,K+-ATPase activity was unchanged after transfer to epithelium of the euryhaline killifish Fundulus freshwater, CA2 mRNA levels decreased, and NHE3 levels heteroclitus. These tissues are functionally similar in increased. Consistent with their functional similarities in seawater, but in freshwater the gills actively absorb Na+ seawater, killifish gills and opercular epithelium expressed – + + + + – but not Cl , whereas the opercular epithelium actively Na ,K -ATPase α1a, Na ,K ,2Cl cotransporter 1 absorbs Cl– but not Na+. These differences in freshwater (NKCC1), cystic fibrosis transmembrane conductance physiology are likely due to differences in absolute levels regulator (CFTR) Cl– channel and the signalling protein of gene expression (measured using real-time PCR), as 14-3-3a at similar absolute levels. Furthermore, NKCC1 several proteins important for Na+ transport, namely and CFTR were suppressed equally in each tissue after Na+,H+-exchanger 2 (NHE2), carbonic anhydrase 2 (CA2), freshwater transfer, and 14-3-3a mRNA increased in both. -

Jlb Smith Institute of Ichthyology
ISSN 0075-2088 J.L.B. SMITH INSTITUTE OF ICHTHYOLOGY GRAHAMSTOWN, SOUTH AFRICA SPECIAL PUBLICATION No. 56 SCIENTIFIC AND COMMON NAMES OF SOUTHERN AFRICAN FRESHWATER FISHES by Paul H. Skelton November 1993 SERIAL PUBLICATIONS o f THE J.L.B. SMITH INSTITUTE OF ICHTHYOLOGY The Institute publishes original research on the systematics, zoogeography, ecology, biology and conservation of fishes. Manuscripts on ancillary subjects (aquaculture, fishery biology, historical ichthyology and archaeology pertaining to fishes) will be considered subject to the availability of publication funds. Two series are produced at irregular intervals: the Special Publication series and the Ichthyological Bulletin series. Acceptance of manuscripts for publication is subject to the approval of reviewers from outside the Institute. Priority is given to papers by staff of the Institute, but manuscripts from outside the Institute will be considered if they are pertinent to the work of the Institute. Colour illustrations can be printed at the expense of the author. Publications of the Institute are available by subscription or in exchange for publi cations of other institutions. Lists of the Institute’s publications are available from the Publications Secretary at the address below. INSTRUCTIONS TO AUTHORS Manuscripts shorter than 30 pages will generally be published in the Special Publications series; longer papers will be considered for the Ichthyological Bulletin series. Please follow the layout and format of a recent Bulletin or Special Publication. Manuscripts must be submitted in duplicate to the Editor, J.L.B. Smith Institute of Ichthyology, Private Bag 1015, Grahamstown 6140, South Africa. The typescript must be double-spaced throughout with 25 mm margins all round. -
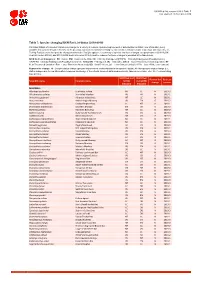
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

The African Turquoise Killifish: a Model for Exploring Vertebrate Aging and Diseases in the Fast Lane
Downloaded from symposium.cshlp.org on June 11, 2016 - Published by Cold Spring Harbor Laboratory Press The African Turquoise Killifish: A Model for Exploring Vertebrate Aging and Diseases in the Fast Lane 1 1,2 ITAMAR HAREL AND ANNE BRUNET 1Department of Genetics, Stanford University, Stanford, California 94305 2Glenn Laboratories for the Biology of Aging at Stanford, Stanford, California 94305 Correspondence: [email protected] Why and how organisms age remains a mystery, and it defines one of the biggest challenges in biology. Aging is also the primary risk factor for many human pathologies, such as cancer, diabetes, cardiovascular diseases, and neurodegenerative diseases. Thus, manipulating the aging rate and potentially postponing the onset of these devastating diseases could have a tremendous impact on human health. Recent studies, relying primarily on nonvertebrate short-lived model systems, have shown the importance of both genetic and environmental factors in modulating the aging rate. However, relatively little is known about aging in vertebrates or what processes may be unique and specific to these complex organisms. Here we discuss how advances in genomics and genome editing have significantly expanded our ability to probe the aging process in a vertebrate system. We highlight recent findings from a naturally short-lived vertebrate, the African turquoise killifish, which provides an attractive platform for exploring mechanisms underlying vertebrate aging and age-related diseases. MODELING AGING IN THE ing vertebrate aging and age-related diseases (Harel et al. LABORATORY 2015). Aging research has greatly benefited from investigating short-lived nonvertebrate models (Fig. 1A), specifically THE AFRICAN TURQUOISE KILLIFISH yeast (Saccharomyces cerevisiae), worm (Caenorhabdi- tis elegans), and fly (Drosophila melanogaster). -

The Neotropical Genus Austrolebias: an Emerging Model of Annual Killifishes Nibia Berois1, Maria J
lopmen ve ta e l B D io & l l o l g e y C Cell & Developmental Biology Berois, et al., Cell Dev Biol 2014, 3:2 ISSN: 2168-9296 DOI: 10.4172/2168-9296.1000136 Review Article Open Access The Neotropical Genus Austrolebias: An Emerging Model of Annual Killifishes Nibia Berois1, Maria J. Arezo1 and Rafael O. de Sá2* 1Departamento de Biologia Celular y Molecular, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay 2Department of Biology, University of Richmond, Richmond, Virginia, USA *Corresponding author: Rafael O. de Sá, Department of Biology, University of Richmond, Richmond, Virginia, USA, Tel: 804-2898542; Fax: 804-289-8233; E-mail: [email protected] Rec date: Apr 17, 2014; Acc date: May 24, 2014; Pub date: May 27, 2014 Copyright: © 2014 Rafael O. de Sá, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Annual fishes are found in both Africa and South America occupying ephemeral ponds that dried seasonally. Neotropical annual fishes are members of the family Rivulidae that consist of both annual and non-annual fishes. Annual species are characterized by a prolonged embryonic development and a relatively short adult life. Males and females show striking sexual dimorphisms, complex courtship, and mating behaviors. The prolonged embryonic stage has several traits including embryos that are resistant to desiccation and undergo up to three reversible developmental arrests until hatching. These unique developmental adaptations are closely related to the annual fish life cycle and are the key to the survival of the species. -

Draft Risk Assessment Report Nothobranchius Furzeri (Turquoise
APPLICATION TO AMEND THE LIST OF SPECIMENS SUITABLE FOR LIVE IMPORT Draft Risk Assessment Report 1. Taxonomy of the species a. Family name: Aplocheilidae b. Genus name: Nothobranchius c. Species: N. furzeri d. Subspecies: No e. Taxonomic Reference: http://www.uniprot.org/taxonomy/105023 f. Common Names: Turquoise killifish g. Is the species a genetically-modified organism (GMO)? No 2. Status of the species under CITES Species is not listed in the CITES Appendices. 3. Ecology of the species a. Longevity: what is the average lifespan of the species in the wild and in captivity? The average lifespan of the species in captivity is 13 weeks (1). Wild derived N. furzeri from three different habitats showed a maximum lifespan of 25-32 weeks (1). b. What is the maximum length and weight that the species attains? Provide information on the size and weight range for males and females of the species. As per the original formal description of the species, the standard length of N. furzeri from a wild population ranged from 2.5cm to 4.4cm (2). The maximum length the species can attain is 7cm (3). The mean maximal body weight reported for males is 3.8 ± 1.1 g (range: 0.5 – 6.7 g) and 2.2 ± 0.6 g (range: 0.2 - 3.6 g) for females (4). Growth rates of Nothobranchius in the wild have been reported to be similar to those in captivity (5). c. Discuss the identification of the individuals in this species, including if the sexes of the species are readily distinguishable, and if the species is difficult to distinguish from other species. -

01 Astyanax Final Version.Indd
Vertebrate Zoology 59 (1) 2009 31 31 – 40 © Museum für Tierkunde Dresden, ISSN 1864-5755, 29.05.2009 Osteology of the African annual killifi sh genus Callopanchax (Teleostei: Cyprinodontiformes: Nothobranchiidae) and phylogenetic implications WILSON J. E. M. COSTA Laboratório de Ictiologia Geral e Aplicada, Departamento de Zoologia, Universidade Federal do Rio de Janeiro, Caixa Postal 68049, CEP 21944-970, Rio de Janeiro, Brazil E-mail: wcosta(at)acd.ufrj.br Received on May 5, 2008, accepted on October 6, 2008. Published online at www.vertebrate-zoology.de on May 15, 2009. > Abstract Osteological structures of Callopanchax are fi rst described and illustrated. Twenty-six characters derived from comparisons of osseous structures among some aplocheiloid fi shes provided evidence supporting hypotheses of relationships among three western African genera (Callopanchax, Scriptaphyosemion and Archiaphyosemion), as proposed in recent molecular analysis. The clade comprising Callopanchax, Scriptaphyosemion and Archiaphyosemion is supported by a laterally displaced antero-proximal process of the fourth ceratobranchial. The sister group relationship between Callopanchax and Scriptaphyosemion is supported by a constriction on the posterior portion of the parasphenoid, an anterior expansion of the hyomandibula, a rectangular basihyal cartilage, an anterior pointed process on the fi rst vertebra, and a long ventrally directed hemal prezygapophysis on the preural centrum 2. Monophyly of Callopanchax is supported by a convexity on the dorsal margin of the opercle, a long interarcual cartilage, and long neural prezygapophyses on the anterior caudal vertebrae. > Key words Killifi shes, Callopanchax, Africa, Osteology, Annual fi shes. Introduction COSTA, 1998a, 2004) and among genera and species of the Rivulidae (e. g., COSTA, 1998b, 2005, 2006a, b).