Ploidy Levels and Relative Genome Sizes of Species, Hybrids, And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
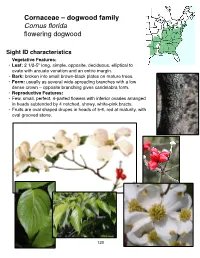
Cornaceae – Dogwood Family Cornus Florida Flowering Dogwood
Cornaceae – dogwood family Cornus florida flowering dogwood Sight ID characteristics Vegetative Features: • Leaf: 2 1/2-5" long, simple, opposite, deciduous, elliptical to ovate with arcuate venation and an entire margin. • Bark: broken into small brown-black plates on mature trees. • Form: usually as several wide-spreading branches with a low dense crown – opposite branching gives candelabra form. • Reproductive Features: • Few, small, perfect, 4-parted flowers with inferior ovaries arranged in heads subtended by 4 notched, showy, white-pink bracts. • Fruits are oval shaped drupes in heads of 5-6, red at maturity, with oval grooved stone. 123 NOTES AND SKETCHES 124 Cornaceae – dogwood family Cornus nuttallii Pacific dogwood Sight ID characteristics Vegetative Features: • Leaf: 2 1/2-4 1/2" long, simple, opposite, deciduous, ovate- elliptical with arcuate venation, margin may be sparsely toothed or entire. • Bark: dark and broken into small plates at maturity. • Form: straight trunk and narrow crown in forested conditions, many-trunked and bushy in open. • Reproductive Features: • Many yellowish-green, small, perfect, 4-parted flowers with inferior ovaries arranged in dense in heads, subtended by 4-7 showy white- pink, petal-like bracts - not notched at the apex. • Fruits are drupes in heads of 30-40, red at maturity and they have smooth stones. 125 NOTES AND SKETCHES 126 Cornaceae – dogwood family Cornus sericea red-osier dogwood Sight ID characteristics Vegetative Features: • Leaf: 2-4" long, simple, opposite, deciduous and somewhat narrow ovate-lanceolate with entire margin. • Twig: bright red, sometimes green splotched with red, white pith. • Bark: red to green with numerous lenticels; later developing larger cracks and splits and turning light brown. -

Likely to Have Habitat Within Iras That ALLOW Road
Item 3a - Sensitive Species National Master List By Region and Species Group Not likely to have habitat within IRAs Not likely to have Federal Likely to have habitat that DO NOT ALLOW habitat within IRAs Candidate within IRAs that DO Likely to have habitat road (re)construction that ALLOW road Forest Service Species Under NOT ALLOW road within IRAs that ALLOW but could be (re)construction but Species Scientific Name Common Name Species Group Region ESA (re)construction? road (re)construction? affected? could be affected? Bufo boreas boreas Boreal Western Toad Amphibian 1 No Yes Yes No No Plethodon vandykei idahoensis Coeur D'Alene Salamander Amphibian 1 No Yes Yes No No Rana pipiens Northern Leopard Frog Amphibian 1 No Yes Yes No No Accipiter gentilis Northern Goshawk Bird 1 No Yes Yes No No Ammodramus bairdii Baird's Sparrow Bird 1 No No Yes No No Anthus spragueii Sprague's Pipit Bird 1 No No Yes No No Centrocercus urophasianus Sage Grouse Bird 1 No Yes Yes No No Cygnus buccinator Trumpeter Swan Bird 1 No Yes Yes No No Falco peregrinus anatum American Peregrine Falcon Bird 1 No Yes Yes No No Gavia immer Common Loon Bird 1 No Yes Yes No No Histrionicus histrionicus Harlequin Duck Bird 1 No Yes Yes No No Lanius ludovicianus Loggerhead Shrike Bird 1 No Yes Yes No No Oreortyx pictus Mountain Quail Bird 1 No Yes Yes No No Otus flammeolus Flammulated Owl Bird 1 No Yes Yes No No Picoides albolarvatus White-Headed Woodpecker Bird 1 No Yes Yes No No Picoides arcticus Black-Backed Woodpecker Bird 1 No Yes Yes No No Speotyto cunicularia Burrowing -

NATIVE PLANT FIELD GUIDE Revised March 2012
NATIVE PLANT FIELD GUIDE Revised March 2012 Hansen's Northwest Native Plant Database www.nwplants.com Foreword Once upon a time, there was a very kind older gentleman who loved native plants. He lived in the Pacific northwest, so plants from this area were his focus. As a young lad, his grandfather showed him flowers and bushes and trees, the sweet taste of huckleberries and strawberries, the smell of Giant Sequoias, Incense Cedars, Junipers, pines and fir trees. He saw hummingbirds poking Honeysuckles and Columbines. He wandered the woods and discovered trillium. When he grew up, he still loved native plants--they were his passion. He built a garden of natives and then built a nursery so he could grow lots of plants and teach gardeners about them. He knew that alien plants and hybrids did not usually live peacefully with natives. In fact, most of them are fierce enemies, not well behaved, indeed, they crowd out and overtake natives. He wanted to share his information so he built a website. It had a front page, a page of plants on sale, and a page on how to plant natives. But he wanted more, lots more. So he asked for help. I volunteered and he began describing what he wanted his website to do, what it should look like, what it should say. He shared with me his dream of making his website so full of information, so inspiring, so educational that it would be the most important source of native plant lore on the internet, serving the entire world. -

Open As a Single Document
Cercis: The Redbuds by KENNETH R. ROBERTSON One of the few woody plants native to eastern North America that is widely planted as an ornamental is the eastern redbud, Cercis canadensis. This plant belongs to a genus of about eight species that is of interest to plant geographers because of its occurrence in four widely separated areas - the eastern United States southwestward to Mexico; western North America; south- ern and eastern Europe and western Asia; and eastern Asia. Cercis is a very distinctive genus in the Caesalpinia subfamily of the legume family (Leguminosae subfamily Caesalpinioi- - deae). Because the apparently simple heart-shaped leaves are actually derived from the fusion of two leaflets of an evenly pinnately compound leaf, Cercis is thought to be related to -~auhinic~, which includes the so-called orchid-trees c$~~ cultivated in tropical regions. The leaves of Bauhinia are usu- ally two-lobed with an apical notch and are made up of clearly ~ two partly fused leaflets. The eastern redbud is more important in the garden than most other spring flowering trees because the flower buds, as well as the open flowers, are colorful, and the total ornamental season continues for two to three weeks. In winter a small bud is found just above each of the leaf scars that occur along the twigs of the previous year’s growth; there are also clusters of winter buds on older branches and on the tree trunks (Figure 3). In early spring these winter buds enlarge (with the excep- tion of those at the tips of the branches) and soon open to re- veal clusters of flower buds. -

Botanischer Garten Der Universität Tübingen
Botanischer Garten der Universität Tübingen 1974 – 2008 2 System FRANZ OBERWINKLER Emeritus für Spezielle Botanik und Mykologie Ehemaliger Direktor des Botanischen Gartens 2016 2016 zur Erinnerung an LEONHART FUCHS (1501-1566), 450. Todesjahr 40 Jahre Alpenpflanzen-Lehrpfad am Iseler, Oberjoch, ab 1976 20 Jahre Förderkreis Botanischer Garten der Universität Tübingen, ab 1996 für alle, die im Garten gearbeitet und nachgedacht haben 2 Inhalt Vorwort ...................................................................................................................................... 8 Baupläne und Funktionen der Blüten ......................................................................................... 9 Hierarchie der Taxa .................................................................................................................. 13 Systeme der Bedecktsamer, Magnoliophytina ......................................................................... 15 Das System von ANTOINE-LAURENT DE JUSSIEU ................................................................. 16 Das System von AUGUST EICHLER ....................................................................................... 17 Das System von ADOLF ENGLER .......................................................................................... 19 Das System von ARMEN TAKHTAJAN ................................................................................... 21 Das System nach molekularen Phylogenien ........................................................................ 22 -

Cornus Florida
Cornus florida Family: Cornaceae Flowering Dogwood The genus Cornus contains about 40 species which grow in the northern temperate regions of the world. The name cornus is derived from the Latin name of the type species Cornus mas L., Cornelian-cherry of Europe, from the word for horn (cornu), referring to the hardness of the wood. Cornus alternifolia- Alternate Leaf Dogwood, Blue Dogwood, Green-Osier, Pagoda, Pagoda Cornel, Pagoda Dogwood, Pigeonberry, Purple Dogwood, Umbrella-tree Cornus drummondii-Roughleaf Dogwood, Rough-leaved Dogwood Cornus florida- Arrowwood, Boxwood, Bunchberry, Cornel, Dogwood (used bark to treat dog's mange), False Boxwood, Florida Dogwood, Flowering Dogwood, White Cornel Cornus glabrata-Brown Dogwood, Flowering Dogwood, Mountain Dogwood, Pacific Dogwood, Smooth Dogwood, Western Flowering Dogwood Cornus nuttallii-California Dogwood, Flowering Dogwood, Mountain Dogwood, Pacific Dogwood, Western Dogwood, Western Flowering Dogwood Cornus occidentalis-Western Dogwood Cornus racemosa-Blue-fruit Dogwood, Gray Dogwood, Stiffcornel, Stiff Cornel Dogwood, Stiff Dogwood, Swamp Dogwood Cornus rugosa-Roundleaf Dogwood Cornus sessilis-Blackfruit Dogwood, Miners Dogwood Cornus stolonifera-American Dogwood, California Dogwood, Creek Dogwood, Kinnikinnik, Red Dogwood, Red-Osier Dogwood, Red-panicled Dogwood, Redstem Dogwood, Squawbush, Western Dogwood Cornus stricta-Bluefruit Dogwood, Stiffcornel, Stiffcornel Dogwood, Swamp Dogwood The following is for Flowering Dogwood: Distribution North America, from Maine to New York, Ontario, Michigan, Illinois and Missouri south to Kansas, Oklahoma and Texas east to Florida. The Tree Flowering dogwood is well known for its white flower clusters with large white bracts opening in the spring. The fall foliage is bright red. It is a slow growing tree which attains a height of 40 feet and a diameter of 16 inches. -

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -

Cornus Nuttallii 'Monarch'
http://vdberk.demo-account.nl/trees/cornus-nuttallii-monarch/ Cornaceae Cornus Cornus nuttallii 'Monarch' Height 6 - 8 m Crown wide conical , half-open crown, capricious growing Bark and branches red brown to grey, flaking in small plates Leaf wide ovate to oval, green, 6 - 12 cm Attractive autumn colour yellow, orange, red Flowers green yellow in heads, inconspicuous, bracts white, May Fruits ovoid berry-like stone-fruit, Ø 1 cm, bright red Spines/thorns none Toxicity non-toxic (usually) Soil type humus rich content and moisture-retentive Paving tolerates no paving Winter hardiness 7a (-17,7 to -15,0 °C) Wind resistance moderate Light requirement suitable for shadow Fauna tree valuable for butterflies, provides food for birds Application parks, tree containers, theme parks, cemeteries, roof gardens, large gardens, small gardens, patio gardens Type/shape clearstem tree, feathered tree, specimen shrub Origin A. van der Bom, Oudenbosch (NL), before 1970 This cultivar 'Monarch' has an upright habit with a good upright central leader. Therefore it is better suited as a tree, which distinguishes it from the species. Young twigs are green but turn to brown red quickly. Mature trunks too, are red brown to grey. The green leaf turns yellow to orange red in autumn. The flowers are not showy. However, each head with flowers is surrounded by 6 (sometimes 4 or 8) ovoid, pointed bracts. These turn from cream white to entirely white, sometimes with a pink hue and can become 10 cm. This makes the plant in full bloom very decorative. 'Monarch' flowers profusely. The circa 1 cm large, orange-red fruits appear in early autumn. -

Steciana Doi:10.12657/Steciana.020.004 ISSN 1689-653X
2016, Vol. 20(1): 21–32 Steciana doi:10.12657/steciana.020.004 www.up.poznan.pl/steciana ISSN 1689-653X ANATOMICAL STUDY OF CORNUS MAS L. AND CORNUS OFFICINALIS SEIB. ET ZUCC. (CORNACEAE) ENDOCARPS DURING THEIR DEVELOPMENT MARIA MOROZOWSKA, ILONA WYSAKOWSKA M. Morozowska, I. Wysakowska, Department of Botany, Poznań University of Life Sciences, Wojska Polskiego 71 C, 60-625 Poznań, Poland, e-mail: [email protected], [email protected] (Received: June 17, 2015. Accepted: October 20, 2015) ABSTRACT. Results of anatomical studies on the developing endocarps of Cornus mas and C. officinalis are pre- sented. Formation of an endocarp and anatomical changes in its structure from the flowering stage to fully developed fruits were observed with the use of LM and SEM. In the process of anatomical development of endocarps the formation of two layers, i.e. the inner and the outer endocarp, was observed. Changes in their anatomical structure consisted in a gradual thickening of cell walls and their lignification. The ligni- fication of endocarp cell walls begins in the inner endocarp, it proceeds in the outside parts of the outer endocarp, with an exception of several layers of cells forming the transition zone (circular strand) present on its margin, and finally, almost at the same time, in the rest of the outer endocarp. Cell walls of the cells distinguishing the germinating valves undergo thickening delayed by several days in comparison to other endocarp cells. Thickening of their cell walls starts in cells situated close to the inner endocarp and proceeds to its outer parts, and their lignification is not very intensive. -

Staff Summary for April 15-16, 2020
Item No. 30 STAFF SUMMARY FOR APRIL 15-16, 2020 30. SHASTA SNOW-WREATH CESA PETITION Today’s Item Information ☐ Action ☒ Consider and potentially act on the petition, DFW’s evaluation report, and comments received to determine whether listing Shasta snow-wreath (Neviusia cliftonii) as a threatened or endangered species under the California Endangered Species Act (CESA) may be warranted. Summary of Previous/Future Actions • Received petition Sep 30, 2019 • FGC transmitted petition to DFW Oct 10, 2019 • Published notice of receipt of petition Nov 22, 2020 • Public receipt of petition Dec 11-12, 2019; Sacramento • Received DFW 90-day evaluation report Feb 21, 2020; Sacramento • Today, determine if petitioned action Apr 15-16, 2020; Teleconference may be warranted Background A petition to list Shasta snow-wreath as endangered under CESA was submitted by Kathleen Roche and the California Native Plant Society on Sep 30, 2019 (Exhibit 1). On Oct 10, 2019, FGC staff transmitted the petition to DFW for review. A notice of receipt of petition was published in the California Regulatory Notice Register on Nov 22, 2019. California Fish and Game Code Section 2073.5 requires that DFW evaluate the petition and submit to FGC a written evaluation with a recommendation, which was received at FGC’s Feb 21, 2020 meeting. The evaluation report (Exhibit 2) delineates each of the categories of information required for a petition, evaluates the sufficiency of the available scientific information for each of the required components, and incorporates additional relevant information that DFW possessed or received during the review period. Today’s agenda item follows the public release and review period of the evaluation report prior to FGC action, as required in Fish and Game Code Section 2074. -

Native Plant List CITY of OREGON CITY 320 Warner Milne Road , P.O
Native Plant List CITY OF OREGON CITY 320 Warner Milne Road , P.O. Box 3040, Oregon City, OR 97045 Phone: (503) 657-0891, Fax: (503) 657-7892 Scientific Name Common Name Habitat Type Wetland Riparian Forest Oak F. Slope Thicket Grass Rocky Wood TREES AND ARBORESCENT SHRUBS Abies grandis Grand Fir X X X X Acer circinatumAS Vine Maple X X X Acer macrophyllum Big-Leaf Maple X X Alnus rubra Red Alder X X X Alnus sinuata Sitka Alder X Arbutus menziesii Madrone X Cornus nuttallii Western Flowering XX Dogwood Cornus sericia ssp. sericea Crataegus douglasii var. Black Hawthorn (wetland XX douglasii form) Crataegus suksdorfii Black Hawthorn (upland XXX XX form) Fraxinus latifolia Oregon Ash X X Holodiscus discolor Oceanspray Malus fuscaAS Western Crabapple X X X Pinus ponderosa Ponderosa Pine X X Populus balsamifera ssp. Black Cottonwood X X Trichocarpa Populus tremuloides Quaking Aspen X X Prunus emarginata Bitter Cherry X X X Prunus virginianaAS Common Chokecherry X X X Pseudotsuga menziesii Douglas Fir X X Pyrus (see Malus) Quercus garryana Garry Oak X X X Quercus garryana Oregon White Oak Rhamnus purshiana Cascara X X X Salix fluviatilisAS Columbia River Willow X X Salix geyeriana Geyer Willow X Salix hookerianaAS Piper's Willow X X Salix lucida ssp. lasiandra Pacific Willow X X Salix rigida var. macrogemma Rigid Willow X X Salix scouleriana Scouler Willow X X X Salix sessilifoliaAS Soft-Leafed Willow X X Salix sitchensisAS Sitka Willow X X Salix spp.* Willows Sambucus spp.* Elderberries Spiraea douglasii Douglas's Spiraea Taxus brevifolia Pacific Yew X X X Thuja plicata Western Red Cedar X X X X Tsuga heterophylla Western Hemlock X X X Scientific Name Common Name Habitat Type Wetland Riparian Forest Oak F. -

Seed Collection and Conservation of Cornus Nuttallii (Pacific Dogwood), a Region 1 Sensitive Species
SEED COLLECTION AND CONSERVATION OF CORNUS NUTTALLII (PACIFIC DOGWOOD), A REGION 1 SENSITIVE SPECIES by Christine C. Lorain Natural Heritage Section Nongame/Endangered Wildlife Program Bureau of Wildlife November 1990 Idaho Department of Fish and Game 600 South Walnut, P.O. Box 25 Boise, Idaho 83707 Jerry M. Conley, Director Cooperative Challenge Cost Share Project Clearwater and Nez Perce National Forests Idaho Department of Fish and Game Purchase Order No. 43-0276-0-0317 (CNF) Purchase Order No. 43-0252-0-0293 (NPNF) ABSTRACT Seed of Cornus nuttallii (Pacific dogwood) was collected and preserved from the Clearwater and Nez Perce National Forests during the 1990 field season. This project was conducted by the Idaho Department of Fish and Game's Natural Heritage Program as a cooperative Challenge Cost Share venture between the Department and the Clearwater and Nez Perce National Forests. Pacific dogwood, a Region 1 Sensitive Plant Species, is a Pacific coastal disjunct to northern Idaho. The principal distribution of this species is west of the Cascade/Sierra crest from southwestern British Columbia to southern California. A single isolated population occurs some 300 miles inland along the confluence of the Lochsa and Selway Rivers on lands administered by the Nez Perce and Clearwater National Forests. In the late 1980's, a severe population decline caused by a root disease was noted within the disjunct population. Due to this serious threat, a comprehensive conservation strategy for Pacific dogwood in Idaho was initiated. Part of this strategy concentrated on preserving this unique disjunct population and its potentially valuable (including commercial) gene pool through seed collection and long-term storage.