Low Divergence Among Natural Populations of Cornus Kousa Subsp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
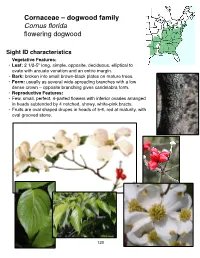
Cornaceae – Dogwood Family Cornus Florida Flowering Dogwood
Cornaceae – dogwood family Cornus florida flowering dogwood Sight ID characteristics Vegetative Features: • Leaf: 2 1/2-5" long, simple, opposite, deciduous, elliptical to ovate with arcuate venation and an entire margin. • Bark: broken into small brown-black plates on mature trees. • Form: usually as several wide-spreading branches with a low dense crown – opposite branching gives candelabra form. • Reproductive Features: • Few, small, perfect, 4-parted flowers with inferior ovaries arranged in heads subtended by 4 notched, showy, white-pink bracts. • Fruits are oval shaped drupes in heads of 5-6, red at maturity, with oval grooved stone. 123 NOTES AND SKETCHES 124 Cornaceae – dogwood family Cornus nuttallii Pacific dogwood Sight ID characteristics Vegetative Features: • Leaf: 2 1/2-4 1/2" long, simple, opposite, deciduous, ovate- elliptical with arcuate venation, margin may be sparsely toothed or entire. • Bark: dark and broken into small plates at maturity. • Form: straight trunk and narrow crown in forested conditions, many-trunked and bushy in open. • Reproductive Features: • Many yellowish-green, small, perfect, 4-parted flowers with inferior ovaries arranged in dense in heads, subtended by 4-7 showy white- pink, petal-like bracts - not notched at the apex. • Fruits are drupes in heads of 30-40, red at maturity and they have smooth stones. 125 NOTES AND SKETCHES 126 Cornaceae – dogwood family Cornus sericea red-osier dogwood Sight ID characteristics Vegetative Features: • Leaf: 2-4" long, simple, opposite, deciduous and somewhat narrow ovate-lanceolate with entire margin. • Twig: bright red, sometimes green splotched with red, white pith. • Bark: red to green with numerous lenticels; later developing larger cracks and splits and turning light brown. -

Department of Planning and Zoning
Department of Planning and Zoning Subject: Howard County Landscape Manual Updates: Recommended Street Tree List (Appendix B) and Recommended Plant List (Appendix C) - Effective July 1, 2010 To: DLD Review Staff Homebuilders Committee From: Kent Sheubrooks, Acting Chief Division of Land Development Date: July 1, 2010 Purpose: The purpose of this policy memorandum is to update the Recommended Plant Lists presently contained in the Landscape Manual. The plant lists were created for the first edition of the Manual in 1993 before information was available about invasive qualities of certain recommended plants contained in those lists (Norway Maple, Bradford Pear, etc.). Additionally, diseases and pests have made some other plants undesirable (Ash, Austrian Pine, etc.). The Howard County General Plan 2000 and subsequent environmental and community planning publications such as the Route 1 and Route 40 Manuals and the Green Neighborhood Design Guidelines have promoted the desirability of using native plants in landscape plantings. Therefore, this policy seeks to update the Recommended Plant Lists by identifying invasive plant species and disease or pest ridden plants for their removal and prohibition from further planting in Howard County and to add other available native plants which have desirable characteristics for street tree or general landscape use for inclusion on the Recommended Plant Lists. Please note that a comprehensive review of the street tree and landscape tree lists were conducted for the purpose of this update, however, only -

Likely to Have Habitat Within Iras That ALLOW Road
Item 3a - Sensitive Species National Master List By Region and Species Group Not likely to have habitat within IRAs Not likely to have Federal Likely to have habitat that DO NOT ALLOW habitat within IRAs Candidate within IRAs that DO Likely to have habitat road (re)construction that ALLOW road Forest Service Species Under NOT ALLOW road within IRAs that ALLOW but could be (re)construction but Species Scientific Name Common Name Species Group Region ESA (re)construction? road (re)construction? affected? could be affected? Bufo boreas boreas Boreal Western Toad Amphibian 1 No Yes Yes No No Plethodon vandykei idahoensis Coeur D'Alene Salamander Amphibian 1 No Yes Yes No No Rana pipiens Northern Leopard Frog Amphibian 1 No Yes Yes No No Accipiter gentilis Northern Goshawk Bird 1 No Yes Yes No No Ammodramus bairdii Baird's Sparrow Bird 1 No No Yes No No Anthus spragueii Sprague's Pipit Bird 1 No No Yes No No Centrocercus urophasianus Sage Grouse Bird 1 No Yes Yes No No Cygnus buccinator Trumpeter Swan Bird 1 No Yes Yes No No Falco peregrinus anatum American Peregrine Falcon Bird 1 No Yes Yes No No Gavia immer Common Loon Bird 1 No Yes Yes No No Histrionicus histrionicus Harlequin Duck Bird 1 No Yes Yes No No Lanius ludovicianus Loggerhead Shrike Bird 1 No Yes Yes No No Oreortyx pictus Mountain Quail Bird 1 No Yes Yes No No Otus flammeolus Flammulated Owl Bird 1 No Yes Yes No No Picoides albolarvatus White-Headed Woodpecker Bird 1 No Yes Yes No No Picoides arcticus Black-Backed Woodpecker Bird 1 No Yes Yes No No Speotyto cunicularia Burrowing -

Cornus Kousa Kousa Dogwood1 Edward F
Fact Sheet ST-191 November 1993 Cornus kousa Kousa Dogwood1 Edward F. Gilman and Dennis G. Watson2 INTRODUCTION Kousa Dogwood grows 15 to 20 feet tall and has beautiful exfoliating bark, long lasting flowers, good fall color, and attractive fruit (Fig. 1). Branches grow upright when the tree is young, but appear in horizontal layers on mature trees. The crown eventually grows wider than it is tall on many specimens. It would be difficult to use too many Kousa Dogwoods. The white, pointed bracts are produced a month later than Flowering Dogwood and are effective for about a month, sometimes longer. The red fruits are edible and they look like a big round raspberry. Birds devour the fruit quickly. Fall color varies from dull red to maroon. GENERAL INFORMATION Scientific name: Cornus kousa Pronunciation: KOR-nus KOO-suh Common name(s): Kousa Dogwood, Chinese Dogwood, Japanese Dogwood Family: Cornaceae USDA hardiness zones: 5 through 8 (Fig. 2) Figure 1. Young Kousa Dogwood. Origin: not native to North America Uses: container or above-ground planter; near a deck DESCRIPTION or patio; screen; specimen Availability: generally available in many areas within Height: 15 to 20 feet its hardiness range Spread: 15 to 20 feet Crown uniformity: symmetrical canopy with a regular (or smooth) outline, and individuals have more or less identical crown forms Crown shape: round Crown density: dense Growth rate: slow 1. This document is adapted from Fact Sheet ST-191, a series of the Environmental Horticulture Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. -

Breeding Powdery Mildew Resistant Dogwoods and More at Rutgers University© T.J
1 11-B-Molnar-Tom-IPPS-2018 Breeding Powdery Mildew Resistant Dogwoods and More at Rutgers © University T.J. Molnar Department of Plant Biology and Pathology, The School of Environmental and Biological Sciences, Rutgers University, 59 Dudley Road, New Brunswick, New Jersey 08901, USA. Email: [email protected] Keywords: Holly, Ilex sp., Cornus sp., hazelnuts, Corylus sp., Erysiphe pulchra , woody ornamental breeding, clonal propagation, budding, seedlings, molecular tools, novel cultivars INTRODUCTION The Rutgers University Woody Ornamental Breeding Program began in 1960 under the direction of Dr. Elwin Orton. The early focus of the program was the breeding of hollies (Ilex sp.), with work on big-bracted dogwoods (Cornus sp.) starting in the 1970s. The program continues today with the addition of hazelnuts (Corylus sp.) for nut production and ornamentals. Over 40 cultivars have been released since the initiation of the program, and a number have become widely grown in the nursery and landscape trade. A list of releases can be found in Molnar and Capik (2013). The most noteworthy Rutgers introductions, at least from the perspective of plants propagated and sold thus far, are likely the hybrid dogwoods. These were largely the results of crossing Cornus kousa with C. florida to create a series of unique plants (subsequently named Cornus × rutgersensis [Mattera et al., 2015]) that combined traits of both parental species to create attractive, high-value landscape specimens. The hybrids generally exhibit increased vigor over their parental species and better drought tolerance than C. florida. However, their most important attribute is likely their resistance or increased tolerance to 1 2 diseases such as dogwood anthracnose (Discula destructiva) and powdery mildew [(PM), Erysiphe pulchra) (Hibben and Daughtrey, 1998; Ranney et al., 1995)]. -

Cornus Kousa (Japanese Flowering Dogwood)
Cornus kousa (Japanese Flowering Dogwood) Cornus kousa is a small tree or shrub, not usually exceeding 6m. It has a slow to medium growth rate and an upright habit, broadening with age. Cream flowers appear in early summer and last for several weeks, turning a slight pinkish brown as they get older. These are actually the bracts surrounding the true flowers which are inconspicu- ous. Strawberry like fruits are produced which are edible but not overly tasty! The foliage is green and ovate through the summer changing to reds June and purples in autumn. Cornus kousa is more robust than the other flower- ing dogwood, Cornus florida and is also resist- 2011 ant the dogwood anthracnose disease making it a popular alternative. Plant Profile Name: Cornus kousa Common Name: Japanese Flowering Dogwood or Kousa dogwood Family: Cornaceae Height: up to 6m Shape: upright when young but broadens with age Demands: Full sun or partial shade, not tolerant of chalky soils Flowers: Cream in early summer Fruit: 2-3cm, strawberry like fruits Autumn Colour: Foliage colours reds and purples Cornus kousa 4.0-4.5m Deepdale Trees Ltd., Tithe Farm, Hatley Road, Potton, Sandy, Beds. SG19 2DX. Tel: 01767 26 26 36 www.deepdale-trees.co.uk Cornus kousa (Japanese Flowering Dogwood) Yamaboushi or Japanese Flowering Dogwood ヤマボウシ Cornus kousa Cornus kousa Chinensis 4.0-5.0m Cornus kousa supplied to RHS Chelsea Flower Show 2011 2.0-2.5m multistem Autumn colour Homebase – Thomas Hoblyn Deepdale Trees Ltd., Tithe Farm, Hatley Road, Potton, Sandy, Beds. SG19 2DX. Tel: 01767 26 26 36 www.deepdale-trees.co.uk. -

Red Seal Landscape Horticulturist Identify Plants and Plant Requirements I (Nakano)
RED SEAL LANDSCAPE HORTICULTURIST IDENTIFY PLANTS AND PLANT REQUIREMENTS I (NAKANO) Michelle Nakano Kwantlen Polytechnic University Book: Red Seal Landscape Horticulturist Identify Plants and Plant Requirements (Nakano) This text is disseminated via the Open Education Resource (OER) LibreTexts Project (https://LibreTexts.org) and like the hundreds of other texts available within this powerful platform, it freely available for reading, printing and "consuming." Most, but not all, pages in the library have licenses that may allow individuals to make changes, save, and print this book. Carefully consult the applicable license(s) before pursuing such effects. Instructors can adopt existing LibreTexts texts or Remix them to quickly build course-specific resources to meet the needs of their students. Unlike traditional textbooks, LibreTexts’ web based origins allow powerful integration of advanced features and new technologies to support learning. The LibreTexts mission is to unite students, faculty and scholars in a cooperative effort to develop an easy-to-use online platform for the construction, customization, and dissemination of OER content to reduce the burdens of unreasonable textbook costs to our students and society. The LibreTexts project is a multi-institutional collaborative venture to develop the next generation of open-access texts to improve postsecondary education at all levels of higher learning by developing an Open Access Resource environment. The project currently consists of 13 independently operating and interconnected libraries that are constantly being optimized by students, faculty, and outside experts to supplant conventional paper-based books. These free textbook alternatives are organized within a central environment that is both vertically (from advance to basic level) and horizontally (across different fields) integrated. -

Botanischer Garten Der Universität Tübingen
Botanischer Garten der Universität Tübingen 1974 – 2008 2 System FRANZ OBERWINKLER Emeritus für Spezielle Botanik und Mykologie Ehemaliger Direktor des Botanischen Gartens 2016 2016 zur Erinnerung an LEONHART FUCHS (1501-1566), 450. Todesjahr 40 Jahre Alpenpflanzen-Lehrpfad am Iseler, Oberjoch, ab 1976 20 Jahre Förderkreis Botanischer Garten der Universität Tübingen, ab 1996 für alle, die im Garten gearbeitet und nachgedacht haben 2 Inhalt Vorwort ...................................................................................................................................... 8 Baupläne und Funktionen der Blüten ......................................................................................... 9 Hierarchie der Taxa .................................................................................................................. 13 Systeme der Bedecktsamer, Magnoliophytina ......................................................................... 15 Das System von ANTOINE-LAURENT DE JUSSIEU ................................................................. 16 Das System von AUGUST EICHLER ....................................................................................... 17 Das System von ADOLF ENGLER .......................................................................................... 19 Das System von ARMEN TAKHTAJAN ................................................................................... 21 Das System nach molekularen Phylogenien ........................................................................ 22 -

Cornus Florida
Cornus florida Family: Cornaceae Flowering Dogwood The genus Cornus contains about 40 species which grow in the northern temperate regions of the world. The name cornus is derived from the Latin name of the type species Cornus mas L., Cornelian-cherry of Europe, from the word for horn (cornu), referring to the hardness of the wood. Cornus alternifolia- Alternate Leaf Dogwood, Blue Dogwood, Green-Osier, Pagoda, Pagoda Cornel, Pagoda Dogwood, Pigeonberry, Purple Dogwood, Umbrella-tree Cornus drummondii-Roughleaf Dogwood, Rough-leaved Dogwood Cornus florida- Arrowwood, Boxwood, Bunchberry, Cornel, Dogwood (used bark to treat dog's mange), False Boxwood, Florida Dogwood, Flowering Dogwood, White Cornel Cornus glabrata-Brown Dogwood, Flowering Dogwood, Mountain Dogwood, Pacific Dogwood, Smooth Dogwood, Western Flowering Dogwood Cornus nuttallii-California Dogwood, Flowering Dogwood, Mountain Dogwood, Pacific Dogwood, Western Dogwood, Western Flowering Dogwood Cornus occidentalis-Western Dogwood Cornus racemosa-Blue-fruit Dogwood, Gray Dogwood, Stiffcornel, Stiff Cornel Dogwood, Stiff Dogwood, Swamp Dogwood Cornus rugosa-Roundleaf Dogwood Cornus sessilis-Blackfruit Dogwood, Miners Dogwood Cornus stolonifera-American Dogwood, California Dogwood, Creek Dogwood, Kinnikinnik, Red Dogwood, Red-Osier Dogwood, Red-panicled Dogwood, Redstem Dogwood, Squawbush, Western Dogwood Cornus stricta-Bluefruit Dogwood, Stiffcornel, Stiffcornel Dogwood, Swamp Dogwood The following is for Flowering Dogwood: Distribution North America, from Maine to New York, Ontario, Michigan, Illinois and Missouri south to Kansas, Oklahoma and Texas east to Florida. The Tree Flowering dogwood is well known for its white flower clusters with large white bracts opening in the spring. The fall foliage is bright red. It is a slow growing tree which attains a height of 40 feet and a diameter of 16 inches. -

Cornus Nuttallii 'Monarch'
http://vdberk.demo-account.nl/trees/cornus-nuttallii-monarch/ Cornaceae Cornus Cornus nuttallii 'Monarch' Height 6 - 8 m Crown wide conical , half-open crown, capricious growing Bark and branches red brown to grey, flaking in small plates Leaf wide ovate to oval, green, 6 - 12 cm Attractive autumn colour yellow, orange, red Flowers green yellow in heads, inconspicuous, bracts white, May Fruits ovoid berry-like stone-fruit, Ø 1 cm, bright red Spines/thorns none Toxicity non-toxic (usually) Soil type humus rich content and moisture-retentive Paving tolerates no paving Winter hardiness 7a (-17,7 to -15,0 °C) Wind resistance moderate Light requirement suitable for shadow Fauna tree valuable for butterflies, provides food for birds Application parks, tree containers, theme parks, cemeteries, roof gardens, large gardens, small gardens, patio gardens Type/shape clearstem tree, feathered tree, specimen shrub Origin A. van der Bom, Oudenbosch (NL), before 1970 This cultivar 'Monarch' has an upright habit with a good upright central leader. Therefore it is better suited as a tree, which distinguishes it from the species. Young twigs are green but turn to brown red quickly. Mature trunks too, are red brown to grey. The green leaf turns yellow to orange red in autumn. The flowers are not showy. However, each head with flowers is surrounded by 6 (sometimes 4 or 8) ovoid, pointed bracts. These turn from cream white to entirely white, sometimes with a pink hue and can become 10 cm. This makes the plant in full bloom very decorative. 'Monarch' flowers profusely. The circa 1 cm large, orange-red fruits appear in early autumn. -

Morphological and Physiological Responses of Cornus Alba to Salt
HORTSCIENCE 55(2):224–230. 2020. https://doi.org/10.21273/HORTSCI14460-19 and Saha, 2014). Plants under drought stress tend to reduce leaf size, stimulate leaf abscis- sion, enhance root growth, and limit photo- Morphological and Physiological synthesis (Taiz et al., 2015). Some plants can maintain water balance under drought condi- Responses of Cornus alba to Salt tions through osmotic adjustment (Farooq et al., 2008). The fact that drought resistance and Drought Stresses under varies among plant species warrants further investigation to evaluate plant responses to drought conditions and select drought-tolerant Greenhouse Conditions plants for landscape use. Qiang Liu Soil salinity is also a global issue and is College of Life Sciences and Technology, Central South University of caused partially by human activities such as irrigation with poor quality water and poor Forestry and Technology, 498 South Shaoshan Road, Changsha, Hunan soil drainage, which result in excess soluble 410004, China; and Hunan Academy of Forestry, 658 South Shaoshan Road, salts in the soil. It is estimated that 20% of Changsha, Hunan 410004, China the irrigated lands in the world are currently affected by salinity stress (Taiz et al., 2015). Youping Sun Salinity induces a series of metabolic dys- Department of Plants, Soils and Climate, Utah State University, 4820 Old functions in plants, including specific ion Main Hill, Logan, UT 84322 toxicity, nutrient imbalance, decreased pho- tosynthesis, and enzyme dysfunction (Munns James Altland and Tester, 2008). The extent of adverse U.S. Department of Agriculture, Agricultural Research Service, Application impact of salinity on plant physiological Technology Research Unit, 1680 Madison Avenue, Wooster, OH 44691 processes depends on the rate and duration of salinity stress. -

Landscape Plants Rated by Deer Resistance
E271 Bulletin For a comprehensive list of our publications visit www.rce.rutgers.edu Landscape Plants Rated by Deer Resistance Pedro Perdomo, Morris County Agricultural Agent Peter Nitzsche, Morris County Agricultural Agent David Drake, Ph.D., Extension Specialist in Wildlife Management The following is a list of landscape plants rated according to their resistance to deer damage. The list was compiled with input from nursery and landscape professionals, Cooperative Extension personnel, and Master Gardeners in Northern N.J. Realizing that no plant is deer proof, plants in the Rarely Damaged, and Seldom Rarely Damaged categories would be best for landscapes prone to deer damage. Plants Occasionally Severely Damaged and Frequently Severely Damaged are often preferred by deer and should only be planted with additional protection such as the use of fencing, repellents, etc. Success of any of these plants in the landscape will depend on local deer populations and weather conditions. Latin Name Common Name Latin Name Common Name ANNUALS Petroselinum crispum Parsley Salvia Salvia Rarely Damaged Tagetes patula French Marigold Ageratum houstonianum Ageratum Tropaeolum majus Nasturtium Antirrhinum majus Snapdragon Verbena x hybrida Verbena Brugmansia sp. (Datura) Angel’s Trumpet Zinnia sp. Zinnia Calendula sp. Pot Marigold Catharanthus rosea Annual Vinca Occasionally Severely Damaged Centaurea cineraria Dusty Miller Begonia semperflorens Wax Begonia Cleome sp. Spider Flower Coleus sp. Coleus Consolida ambigua Larkspur Cosmos sp. Cosmos Euphorbia marginata Snow-on-the-Mountain Dahlia sp. Dahlia Helichrysum Strawflower Gerbera jamesonii Gerbera Daisy Heliotropium arborescens Heliotrope Helianthus sp. Sunflower Lobularia maritima Sweet Alyssum Impatiens balsamina Balsam, Touch-Me-Not Matricaria sp. False Camomile Impatiens walleriana Impatiens Myosotis sylvatica Forget-Me-Not Ipomea sp.