Synteny-Based Analyses Indicate That Sequence Divergence Is Not The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differentiation-Defective Phenotypes Revealed by Large-Scale Analyses of Human Pluripotent Stem Cells
Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells Michiyo Koyanagi-Aoia,1, Mari Ohnukia,1, Kazutoshi Takahashia, Keisuke Okitaa, Hisashi Nomab, Yuka Sawamuraa, Ito Teramotoa, Megumi Naritaa, Yoshiko Satoa, Tomoko Ichisakaa, Naoki Amanoa, Akira Watanabea, Asuka Morizanea, Yasuhiro Yamadaa,c, Tosiya Satod, Jun Takahashia,e, and Shinya Yamanakaa,f,2 aCenter for iPS Cell Research and Application, eDepartment of Biological Repair, Institute for Frontier Medical Sciences, and cInstitute for Integrated Cell- Material Sciences, Kyoto University, Kyoto 606-8507, Japan; bDepartment of Data Science, The Institute of Statistical Mathematics, Tokyo 190-8562, Japan; dDepartment of Biostatistics, Kyoto University School of Public Health, Kyoto 606-8501, Japan; and fGladstone Institute of Cardiovascular Disease, San Francisco, CA 94158 Contributed by Shinya Yamanaka, October 30, 2013 (sent for review September 13, 2013) We examined the gene expression and DNA methylation of 49 at least three passages. In addition, we analyzed the original somatic human induced pluripotent stem cells (hiPSCs) and 10 human cells, two human embryonic carcinoma cell (hECC) lines (NTera2 embryonic stem cells and found overlapped variations in gene cloneD1 and 2102Ep 4D3), and three cancer cell lines (HepG2, expression and DNA methylation in the two types of human MCF7, and Jurkat). pluripotent stem cell lines. Comparisons of the in vitro neural The mRNA microarray analyses (Fig. 1A) identified 61 probes fi differentiation of 40 hiPSCs and 10 human embryonic stem cells with signi cant differences in expression between hESCs and < showed that seven hiPSC clones retained a significant number of hiPSCs [t test, false discovery rate (FDR) 0.05]. -

IBRAHIM ALDEAILEJ Phd.2013.Pdf
Bangor University DOCTOR OF PHILOSOPHY Identification and functional characterisation of germ line genes in human cancer cells Aldeailej, Ibrahim Award date: 2013 Awarding institution: Bangor University Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 09. Oct. 2021 Bangor University School of Biological Sciences Identification and functional characterisation of germ line genes in human cancer cells Ph.D. Thesis 2013 IBRAHIM ALDEAILEJ Declaration and Consent Details of the Work I hereby agree to deposit the following item in the digital repository maintained by Bangor University and/or in any other repository authorized for use by Bangor University. Author Name: IBRAHIM MOHAMMED ALDEAILEJ Title: Identification and functional characterisation of germ line genes in human cancer cells Supervisor/Department: Dr. Ramsay James McFarlane/ School of Biological Sciences Funding body (if any): kingdom of Saudi Arabia Government Qualification/Degree obtained: Ph.D. -

Arnau Soler2019.Pdf
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Genetic responses to environmental stress underlying major depressive disorder Aleix Arnau Soler Doctor of Philosophy The University of Edinburgh 2019 Declaration I hereby declare that this thesis has been composed by myself and that the work presented within has not been submitted for any other degree or professional qualification. I confirm that the work submitted is my own, except where work which has formed part of jointly-authored publications has been included. My contribution and those of the other authors to this work are indicated below. I confirm that appropriate credit has been given within this thesis where reference has been made to the work of others. I composed this thesis under guidance of Dr. Pippa Thomson. Chapter 2 has been published in PLOS ONE and is attached in the Appendix A, chapter 4 and chapter 5 are published in Translational Psychiatry and are attached in the Appendix C and D, and I expect to submit chapter 6 as a manuscript for publication. -

Systematic Identification and Characterization of Regulatory Elements Derived from Human Endogenous Retroviruses
RESEARCH ARTICLE Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses Jumpei Ito1,2, Ryota Sugimoto1, Hirofumi Nakaoka1,2, Shiro Yamada3, Tetsuaki Kimura1, Takahide Hayano1, Ituro Inoue1,2* 1 Division of Human Genetics, Department of Integrated Genetics, National Institute of Genetics, 1111 Yata, Mishima, Shizuoka, Japan, 2 Department of Genetics, School of Life Science, SOKENDAI (The Graduate a1111111111 University for Advanced Studies), 1111 Yata, Mishima, Shizuoka, Japan, 3 Department of Pediatrics, Tokai a1111111111 University School of Medicine, 143 Shimokasuya, Isehara, Kanagawa, Japan a1111111111 a1111111111 * [email protected] a1111111111 Abstract Human endogenous retroviruses (HERVs) and other long terminal repeat (LTR)-type OPEN ACCESS retrotransposons (HERV/LTRs) have regulatory elements that possibly influence the tran- Citation: Ito J, Sugimoto R, Nakaoka H, Yamada S, scription of host genes. We systematically identified and characterized these regulatory ele- Kimura T, Hayano T, et al. (2017) Systematic ments based on publicly available datasets of ChIP-Seq of 97 transcription factors (TFs) identification and characterization of regulatory provided by ENCODE and Roadmap Epigenomics projects. We determined transcription elements derived from human endogenous retroviruses. PLoS Genet 13(7): e1006883. https:// factor-binding sites (TFBSs) using the ChIP-Seq datasets and identified TFBSs observed doi.org/10.1371/journal.pgen.1006883 on HERV/LTR sequences (HERV-TFBSs). Overall, 794,972 HERV-TFBSs were identified. Editor: CeÂdric Feschotte, University of Utah School Subsequently, we identified ªHERV/LTR-shared regulatory element (HSRE),º defined as a of Medicine, UNITED STATES TF-binding motif in HERV-TFBSs, shared within a substantial fraction of a HERV/LTR type. -

View a Copy of This Licence, Visit
Robertson et al. BMC Biology (2020) 18:103 https://doi.org/10.1186/s12915-020-00826-z RESEARCH ARTICLE Open Access Large-scale discovery of male reproductive tract-specific genes through analysis of RNA-seq datasets Matthew J. Robertson1,2, Katarzyna Kent3,4,5, Nathan Tharp3,4,5, Kaori Nozawa3,5, Laura Dean3,4,5, Michelle Mathew3,4,5, Sandra L. Grimm2,6, Zhifeng Yu3,5, Christine Légaré7,8, Yoshitaka Fujihara3,5,9,10, Masahito Ikawa9, Robert Sullivan7,8, Cristian Coarfa1,2,6*, Martin M. Matzuk1,3,5,6 and Thomas X. Garcia3,4,5* Abstract Background: The development of a safe, effective, reversible, non-hormonal contraceptive method for men has been an ongoing effort for the past few decades. However, despite significant progress on elucidating the function of key proteins involved in reproduction, understanding male reproductive physiology is limited by incomplete information on the genes expressed in reproductive tissues, and no contraceptive targets have so far reached clinical trials. To advance product development, further identification of novel reproductive tract-specific genes leading to potentially druggable protein targets is imperative. Results: In this study, we expand on previous single tissue, single species studies by integrating analysis of publicly available human and mouse RNA-seq datasets whose initial published purpose was not focused on identifying male reproductive tract-specific targets. We also incorporate analysis of additional newly acquired human and mouse testis and epididymis samples to increase the number of targets identified. We detected a combined total of 1178 genes for which no previous evidence of male reproductive tract-specific expression was annotated, many of which are potentially druggable targets. -
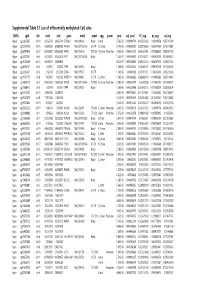
Supplementary Table S1 Kim Et Al.Xlsx
Supplemental Table S1. List of differentially methylated CpG sites DMRs cgid chr start end gene nmid annot cpg prom pval adj. pval FC_avg nc_avg cd_avg Hyper cg12827637 chr14 69256790 69256791 ZFP36L1 NM_004926 Body Island . 5.76E-05 0.000989714 4.465355492 0.06419878 0.287016909 Hyper cg02072495 chr15 60689284 60689285 ANXA2 NM_001136015 5'UTR N_Shore . 2.04E-05 0.000560282 3.808196653 0.056572464 0.215719887 Hyper cg26147845 chr12 132433837 132433838 EP400 NM_015409 TSS1500N_Shore Promoter 3.92E-06 0.000301217 3.643351981 0.077868629 0.283967159 Hyper cg23737061 chr3 60226327 60226328 FHIT NM_001166243 Body . 1.65E-07 0.000239667 3.616168212 0.093920309 0.339893251 Hyper cg18215449 chr12 66089472 66089473 . 4.22E-07 0.000239667 3.565225234 0.064067475 0.228671502 Hyper cg24082121 chr5 672871 672872 TPPP NM_007030 Body . 1.78E-05 0.000525064 3.505843071 0.089095718 0.312606189 Hyper cg23032421 chr3 3152037 3152038 IL5RA NM_175727 5'UTR . 1.04E-05 0.00041338 3.371418731 0.114213604 0.385299026 Hyper cg11737172 chr8 1923051 1923052 KBTBD11 NM_014867 5'UTR S_Shore . 1.38E-05 0.000466865 3.368483202 0.108495063 0.365700645 Hyper cg16886175 chr7 114562062 114562063 MDFIC NM_001166345 TSS200 N_Shore Promoter 7.28E-07 0.000243374 3.35638256 0.101463003 0.340784291 Hyper cg10986412 chr5 672909 672910 TPPP NM_007030 Body . 1.56E-06 0.000255546 3.316242102 0.071981379 0.238939304 Hyper cg02025737 chr15 33384750 33384751 . 3.80E-05 0.000776402 3.312390381 0.13643852 0.452168879 Hyper cg06243675 chr8 11801364 11801365 . 5.23E-05 0.000932639 3.254014485 0.016251643 0.053108482 Hyper cg09577804 chr10 3235531 3235532 . 5.04E-05 0.000912242 3.247042327 0.043496295 0.141459016 Hyper cg23057220 chr19 1356314 1356315 MUM1 NR_024247 TSS200 S_Shore Promoter 3.65E-05 0.000758178 3.204370027 0.14549078 0.466426732 Hyper cg06984883 chr1 1243563 1243564 PUSL1 NM_153339 TSS1500Island Promoter 5.07E-06 0.000323878 3.198831858 0.005047892 0.01636724 Hyper cg15963463 chr1 25253236 25253237 RUNX3 NM_001031680 Body N_Shelf . -

Whole-Genome Sequencing of Finnish Type 1 Diabetic Siblings Discordant for Kidney Disease Reveals DNA Variants Associated with Diabetic Nephropathy
BASIC RESEARCH www.jasn.org Whole-Genome Sequencing of Finnish Type 1 Diabetic Siblings Discordant for Kidney Disease Reveals DNA Variants associated with Diabetic Nephropathy Jing Guo ,1,2 Owen J. L. Rackham ,2 Niina Sandholm ,3,4,5 Bing He ,1 Anne-May Österholm,1,2 Erkka Valo ,3,4,5 Valma Harjutsalo ,3,4,5,6 Carol Forsblom,3,4,5 Iiro Toppila,3,4,5 Maija Parkkonen,3,4,5 Qibin Li,7 Wenjuan Zhu,7 Nathan Harmston ,2,8 Sonia Chothani,2 Miina K. Öhman ,2 Eudora Eng,2 Yang Sun,2 Enrico Petretto ,2,9 Per-Henrik Groop,3,4,5,10 and Karl Tryggvason1,2,11 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background Several genetic susceptibility loci associated with diabetic nephropathy have been documen- ted, but no causative variants implying novel pathogenetic mechanisms have been elucidated. Methods We carried out whole-genome sequencing of a discovery cohort of Finnish siblings with type 1 diabetes who were discordant for the presence (case) or absence (control) of diabetic nephropathy. Con- trols had diabetes without complications for 15–37 years. We analyzed and annotated variants at genome, gene, and single-nucleotide variant levels. We then replicated the associated variants, genes, and regions in a replication cohort from the Finnish Diabetic Nephropathy study that included 3531 unrelated Finns with type 1 diabetes. Results We observed protein-altering variants and an enrichment of variants in regions associated with the presence or absence of diabetic nephropathy. The replication cohort confirmed variants in both regulatory and protein-coding regions. -

Genome-Wide Association Study of Response to Cognitive–Behavioural Therapy in Children with Anxiety Disorders Jonathan R
The British Journal of Psychiatry (2016) 209, 236–243. doi: 10.1192/bjp.bp.115.168229 Genome-wide association study of response to cognitive–behavioural therapy in children with anxiety disorders Jonathan R. I. Coleman, Kathryn J. Lester, Robert Keers, Susanna Roberts, Charles Curtis, Kristian Arendt, Susan Bo¨ gels, Peter Cooper, Cathy Creswell, Tim Dalgleish, Catharina A. Hartman, Einar R. Heiervang, Katrin Ho¨ tzel, Jennifer L. Hudson, Tina In-Albon, Kristen Lavallee, Heidi J. Lyneham, Carla E. Marin, Richard Meiser-Stedman, Talia Morris, Maaike H. Nauta, Ronald M. Rapee, Silvia Schneider, Sophie C. Schneider, Wendy K. Silverman, Mikael Thastum, Kerstin Thirlwall, Polly Waite, Gro Janne Wergeland, Gerome Breen* and Thalia C. Eley* Background Anxiety disorders are common, and cognitive–behavioural suggestive significance (P55610–6) in association with therapy (CBT) is a first-line treatment. Candidate gene studies response post-treatment, and three variants in the 6-month have suggested a genetic basis to treatment response, but follow-up analysis. findings have been inconsistent. Conclusions Aims This is the first genome-wide therapygenetic study. It To perform the first genome-wide association study (GWAS) suggests no common variants of very high effect underlie of psychological treatment response in children with anxiety response to CBT. Future investigations should maximise disorders (n = 980). power to detect single-variant and polygenic effects by using larger, more homogeneous cohorts. Method Presence and severity of anxiety was assessed using semi- Declaration of interest structured interview at baseline, on completion of treatment R.M.R., J.L.H. and H.J.L. are co-authors of the Cool Kids (post-treatment), and 3 to 12 months after treatment program but receive no direct payments. -

The Pluripotent Stem Cell-Specific Transcript ESRG Is Dispensable For
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.25.397935; this version posted November 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 The pluripotent stem cell-specific transcript ESRG is dispensable for human pluripotency 2 3 Kazutoshi Takahashi1, 2, *, Michiko Nakamura1, Megumi Narita1, Akira Watanabe3, Mai Ueda1, Yasuhiro 4 Takashima1, Shinya Yamanaka1, 2, 4 5 6 1Department of Life Science Frontiers, Center for iPS Cell Research and Application, Kyoto University, 7 Kyoto, Japan 8 2Gladstone Institute of Cardiovascular Disease, San Francisco, California, United States of America 9 3Graduate School of Medicine, Kyoto University, Kyoto, Japan 10 4Department of Anatomy, University of California, San Francisco, San Francisco, California, United States 11 of America 12 13 *Corresponding Author 14 E-mail: [email protected] 15 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.11.25.397935; this version posted November 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 16 Abstract 17 Human pluripotent stem cells (PSCs) express human endogenous retrovirus type-H (HERV-H), which 18 exists as more than a thousand copies on the human genome and frequently produces chimeric 19 transcripts as long-non-coding RNAs (lncRNAs) fused with downstream neighbor genes. -

Microrna Regulation of Chondrogenesis in Human Embryonic Stem Cells
MicroRNA Regulation of Chondrogenesis in Human Embryonic Stem Cells 2016 Rosie Sarah Griffiths A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy (PhD) in the Faculty of Biology, Medicine and Health Supervisors: Prof Sue KIMBER and Dr Matthew RONSHAUGEN SCHOOL of BIOLOGICAL SCIENCES Division of Cell Matrix Biology & Regenerative Medicine Contents List of Figures 8 List of Tables 9 List of Abbreviations 11 Abstract 12 Declaration 13 Copyright Statement 14 Acknowledgements 15 Dedication 16 1 Introduction 17 1.1 General Overview . 17 1.2 Articular Cartilage Cell Therapy . 17 1.3 Chondrogenesis . 20 1.3.1 Signaling Pathways Regulating Chondrogenesis . 21 1.3.2 Transcriptional Regulation of Chondrogenesis . 22 1.4 Embryonic Stem Cells . 23 1.4.1 Pluripotency control . 23 1.4.2 Extrinsic Factors Promoting Self-Renewal in ESCs . 24 1.4.3 ESC Characterisation . 25 1.4.4 ESC Culture . 25 1.4.5 Feeder-Free Culture . 26 1.5 MicroRNAs . 26 1.5.1 MicroRNAs in the Genome . 27 1.5.2 Biogenesis . 27 1.5.3 MicroRNA Silencing Mechanism . 30 1.5.4 Target Identification . 30 2 MicroRNA Regulation of Chondrogenesis in Human Embryonic Stem Cells 1.5.5 MicroRNAs in ESCs . 32 1.5.6 MicroRNAs involved in Differentiation . 33 1.5.7 Chondrogenic miRNAs . 34 1.5.8 Manipulating miRNAs . 39 1.6 Next Generation Sequencing and Bioinformatic Analysis . 41 1.6.1 Mapping to miRNAs . 42 1.6.2 Normalisation . 42 1.6.3 Differential Expression of miRNAs . 42 1.6.4 Co-expression Analysis . 43 1.7 Exosomes . -

Title Differentiation-Defective Phenotypes Revealed by Large-Scale
Differentiation-defective phenotypes revealed by large-scale Title analyses of human pluripotent stem cells. Koyanagi-Aoi, Michiyo; Ohnuki, Mari; Takahashi, Kazutoshi; Okita, Keisuke; Noma, Hisashi; Sawamura, Yuka; Teramoto, Author(s) Ito; Narita, Megumi; Sato, Yoshiko; Ichisaka, Tomoko; Amano, Naoki; Watanabe, Akira; Morizane, Asuka; Yamada, Yasuhiro; Sato, Tosiya; Takahashi, Jun; Yamanaka, Shinya Proceedings of the National Academy of Sciences of the Citation United States of America (2013), 110(51): 20569-20574 Issue Date 2013-11-20 URL http://hdl.handle.net/2433/179528 © 2013 National Academy of Sciences; This is not the published version. Please cite only the published version.; この Right 論文は出版社版でありません。引用の際には出版社版を ご確認ご利用ください。 Type Journal Article Textversion author Kyoto University Title Page Classification: BIOLOGICAL SCIENCES/ Cell biology Title: Differentiation defective phenotypes revealed by large scale analyses of human pluripotent stem cells Author affiliation: Michiyo Koyanagi-Aoia,1, Mari Ohnukia,1, Kazutoshi Takahashia, Keisuke Okitaa , Hisashi Nomab, Yuka Sawamuraa, Ito Teramotoa, Megumi Naritaa, Yoshiko Satoa, Tomoko Ichisakaa, Naoki Amanoa, Akira Watanabea, Asuka Morizanea, Yasuhiro Yamadaa,d, Tosiya Satoe, Jun Takahashia,c and Shinya Yamanakaa,f,2 a Center for iPS Cell Research and Application (CiRA), Kyoto University, Kyoto 606-8507, Japan b Department of Data Science, The Institute of Statistical Mathematics, Tokyo 190-8562, Japan c Department of Biological Repair, Institute for Frontier Medical Sciences, Kyoto University, -

Dynamic Regulation of Human Endogenous Retroviruses Mediates Factor-Induced Reprogramming and Differentiation Potential
Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential Mari Ohnukia,1, Koji Tanabea,2, Kenta Sutoua, Ito Teramotoa, Yuka Sawamuraa, Megumi Naritaa, Michiko Nakamuraa, Yumie Tokunagaa, Masahiro Nakamuraa, Akira Watanabea, Shinya Yamanakaa,b,3, and Kazutoshi Takahashia,3 aCenter for iPS Cell Research and Application, Kyoto University, Kyoto 606-8507, Japan; and bGladstone Institute of Cardiovascular Disease, San Francisco, California 94158 Contributed by Shinya Yamanaka, July 14, 2014 (sent for review June 6, 2014; reviewed by Konrad Hochedlinger) Pluripotency can be induced in somatic cells by overexpressing and ESCs, suggesting that HERV-H expression may be a ba- transcription factors, including POU class 5 homeobox 1 (OCT3/4), rometer of pluripotency (5). Species-specific transposable ele- sex determining region Y-box 2 (SOX2), Krüppel-like factor 4 ments, including HERVs, contribute up to 25% of the core (KLF4), and myelocytomatosis oncogene (c-MYC). However, some transcription-factor binding sites in mouse and human pluripo- induced pluripotent stem cells (iPSCs) exhibit defective differenti- tent stem cells, wiring new genes into the core regulatory net- ation and inappropriate maintenance of pluripotency features. work of pluripotency in each species (6). These observations Here we show that dynamic regulation of human endogenous suggest that transposable elements may be important deter- retroviruses (HERVs) is important in the reprogramming process minants of pluripotency.