Differentiation-Defective Phenotypes Revealed by Large-Scale Analyses of Human Pluripotent Stem Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differentiation-Defective Phenotypes Revealed by Large-Scale Analyses of Human Pluripotent Stem Cells
Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells Michiyo Koyanagi-Aoia,1, Mari Ohnukia,1, Kazutoshi Takahashia, Keisuke Okitaa, Hisashi Nomab, Yuka Sawamuraa, Ito Teramotoa, Megumi Naritaa, Yoshiko Satoa, Tomoko Ichisakaa, Naoki Amanoa, Akira Watanabea, Asuka Morizanea, Yasuhiro Yamadaa,c, Tosiya Satod, Jun Takahashia,e, and Shinya Yamanakaa,f,2 aCenter for iPS Cell Research and Application, eDepartment of Biological Repair, Institute for Frontier Medical Sciences, and cInstitute for Integrated Cell- Material Sciences, Kyoto University, Kyoto 606-8507, Japan; bDepartment of Data Science, The Institute of Statistical Mathematics, Tokyo 190-8562, Japan; dDepartment of Biostatistics, Kyoto University School of Public Health, Kyoto 606-8501, Japan; and fGladstone Institute of Cardiovascular Disease, San Francisco, CA 94158 Contributed by Shinya Yamanaka, October 30, 2013 (sent for review September 13, 2013) We examined the gene expression and DNA methylation of 49 at least three passages. In addition, we analyzed the original somatic human induced pluripotent stem cells (hiPSCs) and 10 human cells, two human embryonic carcinoma cell (hECC) lines (NTera2 embryonic stem cells and found overlapped variations in gene cloneD1 and 2102Ep 4D3), and three cancer cell lines (HepG2, expression and DNA methylation in the two types of human MCF7, and Jurkat). pluripotent stem cell lines. Comparisons of the in vitro neural The mRNA microarray analyses (Fig. 1A) identified 61 probes fi differentiation of 40 hiPSCs and 10 human embryonic stem cells with signi cant differences in expression between hESCs and < showed that seven hiPSC clones retained a significant number of hiPSCs [t test, false discovery rate (FDR) 0.05]. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

IBRAHIM ALDEAILEJ Phd.2013.Pdf
Bangor University DOCTOR OF PHILOSOPHY Identification and functional characterisation of germ line genes in human cancer cells Aldeailej, Ibrahim Award date: 2013 Awarding institution: Bangor University Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 09. Oct. 2021 Bangor University School of Biological Sciences Identification and functional characterisation of germ line genes in human cancer cells Ph.D. Thesis 2013 IBRAHIM ALDEAILEJ Declaration and Consent Details of the Work I hereby agree to deposit the following item in the digital repository maintained by Bangor University and/or in any other repository authorized for use by Bangor University. Author Name: IBRAHIM MOHAMMED ALDEAILEJ Title: Identification and functional characterisation of germ line genes in human cancer cells Supervisor/Department: Dr. Ramsay James McFarlane/ School of Biological Sciences Funding body (if any): kingdom of Saudi Arabia Government Qualification/Degree obtained: Ph.D. -

Stroke Genetics and Genomics
Faculdade de Medicina da Universidade de Lisboa Unidade Neurológica de Investigação Clínica PhD Thesis Stroke Genetics and Genomics Tiago Krug Coelho Host Institution: Instituto Gulbenkian de Ciência Supervisor at Instituto Gulbenkian de Ciência: Doctor Sofia Oliveira Supervisor at Faculdade de Medicina da Universidade de Lisboa: Professor José Ferro PhD in Biomedical Sciences Specialization in Neurosciences 2010 Stroke Genetics and Genomics A ciência tem, de facto, um único objectivo: a verdade. Não esgota perfeitamente a sua tarefa se não descobre a causa do todo. Chiara Lubich i Stroke Genetics and Genomics ii Stroke Genetics and Genomics A impressão desta dissertação foi aprovada pela Comissão Coordenadora do Conselho Científico da Faculdade de Medicina de Lisboa em reunião de 28 de Setembro de 2010. iii Stroke Genetics and Genomics iv Stroke Genetics and Genomics As opiniões expressas são da exclusiva responsabilidade do seu autor. v Stroke Genetics and Genomics vi Stroke Genetics and Genomics Abstract ABSTRACT This project presents a comprehensive approach to the identification of new genes that influence the risk for developing stroke. Stroke is the leading cause of death in Portugal and the third leading cause of death in the developed world. It is even more disabling than lethal, and the persistent neurological impairment and physical disability caused by stroke have a very high socioeconomic cost. Moreover, the number of affected individuals is expected to increase with the current aging of the population. Stroke is a “brain attack” cutting off vital blood and oxygen to the brain cells and it is a complex disease resulting from environmental and genetic factors. -

Arnau Soler2019.Pdf
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Genetic responses to environmental stress underlying major depressive disorder Aleix Arnau Soler Doctor of Philosophy The University of Edinburgh 2019 Declaration I hereby declare that this thesis has been composed by myself and that the work presented within has not been submitted for any other degree or professional qualification. I confirm that the work submitted is my own, except where work which has formed part of jointly-authored publications has been included. My contribution and those of the other authors to this work are indicated below. I confirm that appropriate credit has been given within this thesis where reference has been made to the work of others. I composed this thesis under guidance of Dr. Pippa Thomson. Chapter 2 has been published in PLOS ONE and is attached in the Appendix A, chapter 4 and chapter 5 are published in Translational Psychiatry and are attached in the Appendix C and D, and I expect to submit chapter 6 as a manuscript for publication. -

Identification of Biomarkers of Metastatic Disease in Uveal
Identification of biomarkers of metastatic disease in uveal melanoma using proteomic analyses Thesis submitted in accordance with the requirements of the University of Liverpool for the degree of Doctor in Philosophy Martina Angi June 2015 To Mario, the wind beneath my wings 2 Acknowledgments First and foremost, I would like to acknowledge my primary supervisor, Prof. Sarah Coupland, for encouraging me to undergo a PhD and for supporting me in this long journey. I am truly grateful to Dr Helen Kalirai for being the person I could always turn to, for a word of advice on cell culture as much as on parenting skills. I would also like to acknowledge Prof. Bertil Damato for being an inspiration and a mentor; and Dr Sarah Lake and Dr Joseph Slupsky for their precious advice. I would like to thank Dawn, Haleh, Fidan and Fatima for becoming my family away from home, and the other members of the LOORG for the fruitful discussions and lovely cakes. I would like to acknowledge Prof. Heinrich Heimann and the clinical team at LOOC, especially Sisters Hebbar, Johnston, Hachuela and Kaye, for their admirable dedication to UM patients and for their invaluable support to clinical research. I would also like to thank the members of staff in St Paul’s theatre and Simon Biddolph and Anna Ikin in Pathology for their precious help in sample collection. I am grateful to Dr Rosalind Jenkins who guided my first steps in the mysterious word of proteomics, and to Dr Deb Simpsons and Prof. Rob Beynon for showing me its beauty. -

PRODUCT SPECIFICATION Prest Antigen C9orf64 Product
PrEST Antigen C9orf64 Product Datasheet PrEST Antigen PRODUCT SPECIFICATION Product Name PrEST Antigen C9orf64 Product Number APrEST90167 Gene Description chromosome 9 open reading frame 64 Alternative Gene MGC10999 Names Corresponding Anti-C9orf64 (HPA066190) Antibodies Description Recombinant protein fragment of Human C9orf64 Amino Acid Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: SDDLLKKLLKGEMLSYGDRQEVEIRGCSLWCVELIRDCLLELIEQKGEKP NGEINSILLDYYLWDYAHDHREDMKGIPFHRIRCIY Fusion Tag N-terminal His6ABP (ABP = Albumin Binding Protein derived from Streptococcal Protein G) Expression Host E. coli Purification IMAC purification Predicted MW 28 kDa including tags Usage Suitable as control in WB and preadsorption assays using indicated corresponding antibodies. Purity >80% by SDS-PAGE and Coomassie blue staining Buffer PBS and 1M Urea, pH 7.4. Unit Size 100 µl Concentration Lot dependent Storage Upon delivery store at -20°C. Avoid repeated freeze/thaw cycles. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -

Systematic Identification and Characterization of Regulatory Elements Derived from Human Endogenous Retroviruses
RESEARCH ARTICLE Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses Jumpei Ito1,2, Ryota Sugimoto1, Hirofumi Nakaoka1,2, Shiro Yamada3, Tetsuaki Kimura1, Takahide Hayano1, Ituro Inoue1,2* 1 Division of Human Genetics, Department of Integrated Genetics, National Institute of Genetics, 1111 Yata, Mishima, Shizuoka, Japan, 2 Department of Genetics, School of Life Science, SOKENDAI (The Graduate a1111111111 University for Advanced Studies), 1111 Yata, Mishima, Shizuoka, Japan, 3 Department of Pediatrics, Tokai a1111111111 University School of Medicine, 143 Shimokasuya, Isehara, Kanagawa, Japan a1111111111 a1111111111 * [email protected] a1111111111 Abstract Human endogenous retroviruses (HERVs) and other long terminal repeat (LTR)-type OPEN ACCESS retrotransposons (HERV/LTRs) have regulatory elements that possibly influence the tran- Citation: Ito J, Sugimoto R, Nakaoka H, Yamada S, scription of host genes. We systematically identified and characterized these regulatory ele- Kimura T, Hayano T, et al. (2017) Systematic ments based on publicly available datasets of ChIP-Seq of 97 transcription factors (TFs) identification and characterization of regulatory provided by ENCODE and Roadmap Epigenomics projects. We determined transcription elements derived from human endogenous retroviruses. PLoS Genet 13(7): e1006883. https:// factor-binding sites (TFBSs) using the ChIP-Seq datasets and identified TFBSs observed doi.org/10.1371/journal.pgen.1006883 on HERV/LTR sequences (HERV-TFBSs). Overall, 794,972 HERV-TFBSs were identified. Editor: CeÂdric Feschotte, University of Utah School Subsequently, we identified ªHERV/LTR-shared regulatory element (HSRE),º defined as a of Medicine, UNITED STATES TF-binding motif in HERV-TFBSs, shared within a substantial fraction of a HERV/LTR type. -

(12) Patent Application Publication (10) Pub. No.: US 2013/0022974 A1 Chinnaiyan Et Al
US 2013 0022974A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2013/0022974 A1 Chinnaiyan et al. (43) Pub. Date: Jan. 24, 2013 (54) DNA METHYLATION PROFILES IN CANCER Publication Classification (75) Inventors: Arul M. Chinnaiyan, Plymouth, MI (51) Int. Cl. (US); Mohan Saravana Dhanasekaran, Ann Arbor, MI (US); Jung Kim, CI2O I/68 (2006.01) Northville, MI (US) (52) U.S. Cl. ...................................................... 435/6.11 (73) Assignee: THE REGENTS OF THE UNIVERSITY OF MICHIGAN, Ann (57) ABSTRACT Arbor, MI (US) (21) Appl. No.: 13/523,545 The present invention relates to compositions and methods (22) Filed: Jun. 14, 2012 for cancer diagnosis, research and therapy, including but not limited to, cancer markers. In particular, the present invention Related U.S. Application Data relates to methylation levels of genes (e.g., in CGI islands of (60) Provisional application No. 61/498,296, filed on Jun. the promoter regions) as diagnostic markers and clinical tar 17, 2011. gets for prostate cancer. Patent Application Publication Jan. 24, 2013 Sheet 1 of 30 US 2013/0022974 A1 Figure 1 A SS 838 s & S&s&&xis&3:38 a. a. a. a. a. a. a. a. a. 888& R is Patent Application Publication Jan. 24, 2013 Sheet 2 of 30 US 2013/0022974 A1 &- 3838; iiirage::c Benign Adja Patent Application Publication Jan. 24, 2013 Sheet 3 of 30 US 2013/0022974 A1 Figure 3 N = 6619 tique TSS Else.” (6077 Unique Reiseg Genes) t S S SS &S .S. SMyy Na3S S Nd: - alie Nic23 Ci (pg islands Cin 5'Shore of CpG islands On 3 Shore of CpG islands On Non-CpG islancis Patent Application Publica US 2013/0022974 A1 ty?un81 Patent Application Publication Jan. -

Larina Carroll Thesis February 9 2018 FINAL for SUBMISSION
Functional Genomics Studies of Atlantic Salmon (Salmo salar) Development, and Sac Fry Responses to Chronic Incremental Hyperthermia By: Larina A. Carroll A thesis submitted to the Department of Biology and the School of Graduate Studies in partial fulfilment of the requirements for the degree of Master of Science Ocean Sciences Centre and Department of Biology, Memorial University of Newfoundland, St. John’s, Newfoundland and Labrador, Canada ABSTRACT Functional genomics techniques, including the 32K cGRASP microarray and real-time quantitative polymerase chain reaction (QPCR), were used to study gene expression in a single Atlantic salmon cohort: 1) during embryonic and early sac fry development; and 2) after sac fry were exposed to chronic incremental hyperthermia (water temperature increased by 1 °C every 24 hours, from 7.4 to 21.4 °C). The first study focused on the transcript expression of four microarray-identified paralogous γM-crystallin genes as well as four paralogous α-sHSP genes. The four α-sHSPs, as well as cryGM4-like and cryGM3-like, were shown by QPCR as higher expressed at hatch or post-hatch stage relative to eye-up stage (although the degree of induction varied between paralogues). This research provides possible evidence of divergent transcript expression (i.e., regulation) of duplicated genes, suggesting that some of the paralogues studied may have diverged functionally. These results provide insight into the evolutionary relationships between these genes, and may provide evidence of neofunctionalization following GD events. The second study focused on four microarray-identified genes of interest (trypsin-1 precursor, chymotrypsin b, ferritin middle subunit, and ubiquitin) as well as the four paralogous α-sHSP genes. -

View a Copy of This Licence, Visit
Robertson et al. BMC Biology (2020) 18:103 https://doi.org/10.1186/s12915-020-00826-z RESEARCH ARTICLE Open Access Large-scale discovery of male reproductive tract-specific genes through analysis of RNA-seq datasets Matthew J. Robertson1,2, Katarzyna Kent3,4,5, Nathan Tharp3,4,5, Kaori Nozawa3,5, Laura Dean3,4,5, Michelle Mathew3,4,5, Sandra L. Grimm2,6, Zhifeng Yu3,5, Christine Légaré7,8, Yoshitaka Fujihara3,5,9,10, Masahito Ikawa9, Robert Sullivan7,8, Cristian Coarfa1,2,6*, Martin M. Matzuk1,3,5,6 and Thomas X. Garcia3,4,5* Abstract Background: The development of a safe, effective, reversible, non-hormonal contraceptive method for men has been an ongoing effort for the past few decades. However, despite significant progress on elucidating the function of key proteins involved in reproduction, understanding male reproductive physiology is limited by incomplete information on the genes expressed in reproductive tissues, and no contraceptive targets have so far reached clinical trials. To advance product development, further identification of novel reproductive tract-specific genes leading to potentially druggable protein targets is imperative. Results: In this study, we expand on previous single tissue, single species studies by integrating analysis of publicly available human and mouse RNA-seq datasets whose initial published purpose was not focused on identifying male reproductive tract-specific targets. We also incorporate analysis of additional newly acquired human and mouse testis and epididymis samples to increase the number of targets identified. We detected a combined total of 1178 genes for which no previous evidence of male reproductive tract-specific expression was annotated, many of which are potentially druggable targets. -
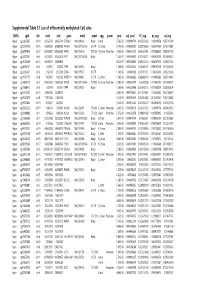
Supplementary Table S1 Kim Et Al.Xlsx
Supplemental Table S1. List of differentially methylated CpG sites DMRs cgid chr start end gene nmid annot cpg prom pval adj. pval FC_avg nc_avg cd_avg Hyper cg12827637 chr14 69256790 69256791 ZFP36L1 NM_004926 Body Island . 5.76E-05 0.000989714 4.465355492 0.06419878 0.287016909 Hyper cg02072495 chr15 60689284 60689285 ANXA2 NM_001136015 5'UTR N_Shore . 2.04E-05 0.000560282 3.808196653 0.056572464 0.215719887 Hyper cg26147845 chr12 132433837 132433838 EP400 NM_015409 TSS1500N_Shore Promoter 3.92E-06 0.000301217 3.643351981 0.077868629 0.283967159 Hyper cg23737061 chr3 60226327 60226328 FHIT NM_001166243 Body . 1.65E-07 0.000239667 3.616168212 0.093920309 0.339893251 Hyper cg18215449 chr12 66089472 66089473 . 4.22E-07 0.000239667 3.565225234 0.064067475 0.228671502 Hyper cg24082121 chr5 672871 672872 TPPP NM_007030 Body . 1.78E-05 0.000525064 3.505843071 0.089095718 0.312606189 Hyper cg23032421 chr3 3152037 3152038 IL5RA NM_175727 5'UTR . 1.04E-05 0.00041338 3.371418731 0.114213604 0.385299026 Hyper cg11737172 chr8 1923051 1923052 KBTBD11 NM_014867 5'UTR S_Shore . 1.38E-05 0.000466865 3.368483202 0.108495063 0.365700645 Hyper cg16886175 chr7 114562062 114562063 MDFIC NM_001166345 TSS200 N_Shore Promoter 7.28E-07 0.000243374 3.35638256 0.101463003 0.340784291 Hyper cg10986412 chr5 672909 672910 TPPP NM_007030 Body . 1.56E-06 0.000255546 3.316242102 0.071981379 0.238939304 Hyper cg02025737 chr15 33384750 33384751 . 3.80E-05 0.000776402 3.312390381 0.13643852 0.452168879 Hyper cg06243675 chr8 11801364 11801365 . 5.23E-05 0.000932639 3.254014485 0.016251643 0.053108482 Hyper cg09577804 chr10 3235531 3235532 . 5.04E-05 0.000912242 3.247042327 0.043496295 0.141459016 Hyper cg23057220 chr19 1356314 1356315 MUM1 NR_024247 TSS200 S_Shore Promoter 3.65E-05 0.000758178 3.204370027 0.14549078 0.466426732 Hyper cg06984883 chr1 1243563 1243564 PUSL1 NM_153339 TSS1500Island Promoter 5.07E-06 0.000323878 3.198831858 0.005047892 0.01636724 Hyper cg15963463 chr1 25253236 25253237 RUNX3 NM_001031680 Body N_Shelf .