(12) Patent Application Publication (10) Pub. No.: US 2009/0035787 A1 Liu Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Summary & Conclusions
Enzymatic functionalization and degradation of natural and synthetic polymers Von der Fakultät für Mathematik, Informatik und Naturwissenschaften der RWTH Aachen University zur Erlangung des akademischen Grades einer Doktorin der Naturwissenschaften genehmigte Dissertation vorgelegt von Shohana Subrin Islam M. Sc. Biotechnologie aus Dhaka, Bangladesch Berichter: Univ. -Prof. Dr. Ulrich Schwaneberg Univ. -Prof. Dr. Lothar Elling Tag der mündlichen Prüfung: 23.01.2019 Diese Dissertation ist auf den Internetseiten der Universitätsbibliothek verfügbar. To my mom & my sister-the two persons in the world who always stand by me Table of content Table of content Table of content _______________________________________________________________ v Publications and patents ________________________________________________________ ix Abstract _____________________________________________________________________ xi 1. General introduction _______________________________________________________ 1 1.1 Enzymatic functionalization of (bio)polymers _______________________________________ 1 1.2 Enzymatic degradation of polymers _______________________________________________ 3 1.3 Protein engineering ____________________________________________________________ 5 1.3.1 Directed evolution of enzymes _________________________________________________________ 6 1.3.2 KnowVolution – Directed Evolution 2.0 __________________________________________________ 9 1.4 Aims of the dissertation _______________________________________________________ 11 2. Engineering of -

Identification of Oxidative Stress-Related Proteins for Predictive Screening of Hepatotoxicity Using a Proteomic Approach
The Journal of Toxicological Sciences, 213 Vol.30, No.3, 213-227, 2005 IDENTIFICATION OF OXIDATIVE STRESS-RELATED PROTEINS FOR PREDICTIVE SCREENING OF HEPATOTOXICITY USING A PROTEOMIC APPROACH Toshinori YAMAMOTO, Rie KIKKAWA, Hiroshi YAMADA and Ikuo HORII Worldwide Safety Sciences, Pfizer Global Research & Development, Nagoya Laboratories, Pfizer Inc., 5-2 Taketoyo, Aichi 470-2393, Japan (Received January 15, 2005; Accepted April 19, 2005) ABSTRACT — We investigated the effects of three hepatotoxicants, acetaminophen (APAP), amio- darone (AD) and tetracycline (TC), on protein expression in primary cultured rat hepatocytes with toxi- coproteomic approach, which is two-dimensional gel electrophoresis (2DE) and mass spectrometry. The objectives of this study were to search for alternative toxicity biomarkers which could be detected with high sensitivity prior to the appearance of morphological changes or alterations of analytical conventional biomarkers. The related proteins in the process of cell degeneration/necrosis such as cell death, lipid metabolism and lipid/carbohydrate metabolism were mainly affected under exposure to APAP, AD and TC, respectively. Among the differentially expressed proteins, several oxidative stress-related proteins were clearly identified after 24-hr exposure, even though they were not affected for 6-hr exposure. They were glutathione peroxidase (GPX) as a down-regulated protein as well as peroxiredoxin 1 (PRX1) and peroxiredoxin 2 (PRX2) as up-regulated proteins, which are known to serve as antioxidative enzymes in cells. These findings suggested that the focused proteins, GPX and PRXs, could be utilized as biomarkers of hepatotoxicity, and they were useful for setting high throughput screening methods to assess hepato- toxicity in the early stage of drug discovery. -

Supporting Information for Proteomics DOI 10.1002/Pmic.200500725
Supporting Information for Proteomics DOI 10.1002/pmic.200500725 Iveta Klouckova, Petra Hrncirova, Yehia Mechref, Randy J. Arnold, Ting-Kai Li, William J. McBride and Milos V. Novotny Changes in liver protein abundance in inbred alcohol-preferring rats due to chronic alcohol exposure, as measured through a proteomics approach ª 2006 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim www.proteomics-journal.com 113 significant proteins GROUP1 GROUP2 ratio of ratio of Coverage average iP1- average iP7- Coverage average iP1- average iP7- Name Abbreviation ssp extract denzity_iP7- ssp extract denzity_iP7- [%] 6" ± "stdev 12" ± "stdev [%] 6" ± "stdev 12" ± "stdev 12/ iP1-6 12/ iP1-6 14-3-3 protein gamma 143G_HUMAN 3209 ex1 24 89 ± 26 53 ± 12 0.59 ± 0.22 2214 ex1 N/A 154 ± 15 136 ± 49 0.89 ± 0.33 Sodium/potassium- transporting ATPase A1A3_RAT 5607 ex1 2 183 ± 119 140 ± 101 0.77 ± 0.75 4727 ex1 2 288 ± 122 100 ± 63 0.35 ± 0.26 alpha-3 chain Aspartate aminotransferase, AATC_RAT 6438 A ex1 8 99 ± 27 67 ± 17 0.67 ± 0.25 6426 ex1 8 348 ± 65 158 ± 61 0.45 ± 0.19 cytoplasmic Acyl-CoA 5328 ex1 14 91 ± 73 48 ± 13 0.53 ± 0.44 5414 ex1 14 486 ± 96 286 ± 99 0.59 ± 0.24 dehydrogenase, ACDB_RAT 7210 ex1 15 181 ± 62 126 ± 32 0.70 ± 0.3 7303 ex1 15 605 ± 125 452 ± 69 0.75 ± 0.19 short/branched chain 5337 B ex1 19 117 ± 60 90 ± 18 0.77 ± 0.43 5429 ex1 19 531 ± 83 273 ± 71 0.52 ± 0.16 Acyl-CoA dehydrogenase, long- ACDL_RAT 5421 C ex1 9 669 ± 151 474 ± 118 0.71 ± 0.24 5417 ex1 N/A 903 ± 267 963 ± 160 1.07 ± 0.36 chain specific Acyl-CoA dehydrogenase, ACDM_RAT 7413 D ex1 -

Structural and Functional Analysis of Heparan Sulfate Sulfotransferases
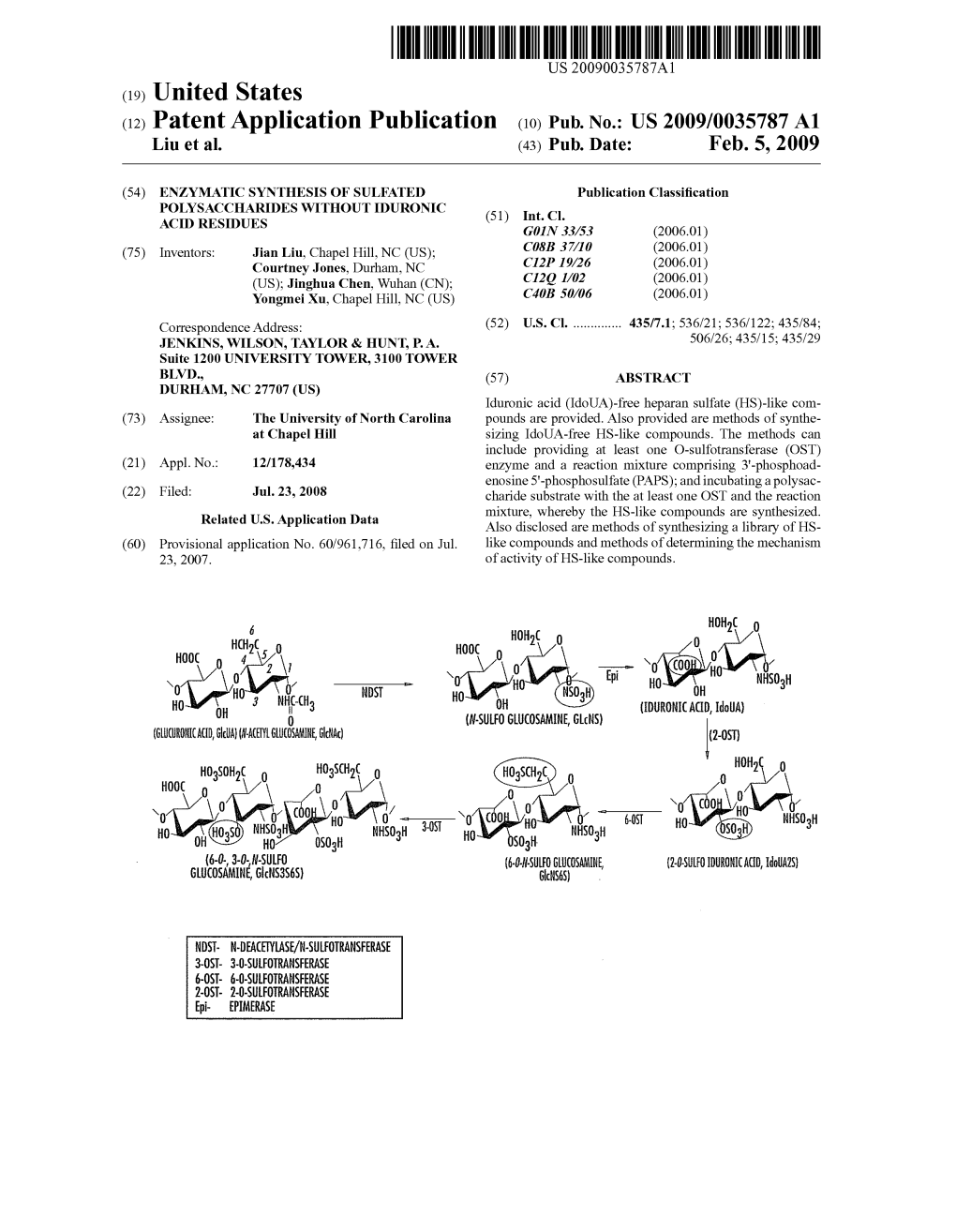
STRUCTURAL AND FUNCTIONAL ANALYSIS OF HEPARAN SULFATE SULFOTRANSFERASES Heather Nicole Bethea A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Pharmaceutical Sciences (Medicinal Chemistry and Natural Products) Chapel Hill 2010 Approved by: Michael Jarstfer, Ph.D. Jian Liu, Ph.D. Lars Pedersen, Ph.D. Scott Singleton, Ph.D. Qisheng Zhang, Ph.D. ABSTRACT HEATHER BETHEA: Structural and Functional Analysis of Heparan Sulfate Sulfotransferases (Under the direction of Jian Liu, Ph.D.) Heparan sulfate (HS), a major polysaccharide component of the vascular system, is involved in regulating a number of functions of the blood vessel wall including blood coagulation, cell differentiation, and the inflammatory response. The wide range of biological functions makes HS an attractive therapeutic target. The long term goal of our research involves utilizing an enzyme-based approach to develop HS-based therapeutics for treating thrombotic diseases, cancer and excessive inflammatory responses. The biosynthesis of HS involves multiple specialized sulfotransferases such as 2-O-sulfotransferase (2OST) and 6-O-sulfotransferase (6OST), which are essential for preparing HS with activities in regulating vascular development and blood coagulation. The substrate specificity of the HS sulfotransferases controls the sulfation patterns of HS, permitting HS to exhibit a specific function, however, limited knowledge regarding the mechanism of these enzymes has hindered our ability to prepare functionally-specific HS. We aim to understand the mechanism of action of these two enzymes in hopes of developing heparin/HS with improved anticoagulant efficacy. In this dissertation, we present successful crystallization of 2OST in complex with 3’- phosphoadenosine 5’-phosphate (PAP). -

X-Ray Fluorescence Analysis Method Röntgenfluoreszenz-Analyseverfahren Procédé D’Analyse Par Rayons X Fluorescents
(19) & (11) EP 2 084 519 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: G01N 23/223 (2006.01) G01T 1/36 (2006.01) 01.08.2012 Bulletin 2012/31 C12Q 1/00 (2006.01) (21) Application number: 07874491.9 (86) International application number: PCT/US2007/021888 (22) Date of filing: 10.10.2007 (87) International publication number: WO 2008/127291 (23.10.2008 Gazette 2008/43) (54) X-RAY FLUORESCENCE ANALYSIS METHOD RÖNTGENFLUORESZENZ-ANALYSEVERFAHREN PROCÉDÉ D’ANALYSE PAR RAYONS X FLUORESCENTS (84) Designated Contracting States: • BURRELL, Anthony, K. AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Los Alamos, NM 87544 (US) HU IE IS IT LI LT LU LV MC MT NL PL PT RO SE SI SK TR (74) Representative: Albrecht, Thomas Kraus & Weisert (30) Priority: 10.10.2006 US 850594 P Patent- und Rechtsanwälte Thomas-Wimmer-Ring 15 (43) Date of publication of application: 80539 München (DE) 05.08.2009 Bulletin 2009/32 (56) References cited: (60) Divisional application: JP-A- 2001 289 802 US-A1- 2003 027 129 12164870.3 US-A1- 2003 027 129 US-A1- 2004 004 183 US-A1- 2004 017 884 US-A1- 2004 017 884 (73) Proprietors: US-A1- 2004 093 526 US-A1- 2004 235 059 • Los Alamos National Security, LLC US-A1- 2004 235 059 US-A1- 2005 011 818 Los Alamos, NM 87545 (US) US-A1- 2005 011 818 US-B1- 6 329 209 • Caldera Pharmaceuticals, INC. US-B2- 6 719 147 Los Alamos, NM 87544 (US) • GOLDIN E M ET AL: "Quantitation of antibody (72) Inventors: binding to cell surface antigens by X-ray • BIRNBAUM, Eva, R. -

Regeneration of PAPS for the Enzymatic Synthesis of Sulfated Oligosaccharides
J. Org. Chem. 2000, 65, 5565-5574 5565 Regeneration of PAPS for the Enzymatic Synthesis of Sulfated Oligosaccharides Michael D. Burkart, Masayuki Izumi, Eli Chapman, Chun-Hung Lin,† and Chi-Huey Wong* Department of Chemistry and The Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037 [email protected] Received February 24, 2000 This paper describes the study of 3′-phosphoadenosine-5′-phosphosulfate (PAPS) regeneration from 3′-phosphoadenosine-5′-phosphate (PAP) for use in practical syntheses of carbohydrate sulfates which are catalyzed by sulfotransferases. Among the regeneration systems, the one with recombinant aryl sulfotransferase proved to be the most practical. This regeneration system was coupled with a sulfotransferase-catalyzed reaction, using a recombinant Nod factor sulfotransferase, for the synthesis of various oligosaccharide sulfates that were further glycosylated using glycosyl- transferases. Sulfated carbohydrates and glycopeptides have gener- modulation of receptor binding.1,8 Drug design for the ated interest due to their roles in specific cell signaling inhibition of these therapeutically interesting enzymes and recognition events of both normal and disease has quickly followed sulfotransferase discovery and will processes,1 such as chronic inflammation, cancer me- gain increasing importance as we have easy access to this tastasis, cartilage formation, and hormone regulation. class of molecules to study their biological roles more Recent studies -

(Sult1e1) Expression and Function in Mcf10a-Series Breast Epithelial Cells
Wayne State University Wayne State University Dissertations 1-1-2011 Estrogen sulfotransferase (sult1e1) expression and function in mcf10a-series breast epithelial cells: role as a modifier of breast carcinogenesis and regulation by proliferation state Jiaqi Fu Wayne State University, Follow this and additional works at: http://digitalcommons.wayne.edu/oa_dissertations Part of the Pathology Commons, and the Toxicology Commons Recommended Citation Fu, Jiaqi, "Estrogen sulfotransferase (sult1e1) expression and function in mcf10a-series breast epithelial cells: role as a modifier of breast carcinogenesis and regulation by proliferation state" (2011). Wayne State University Dissertations. Paper 308. This Open Access Dissertation is brought to you for free and open access by DigitalCommons@WayneState. It has been accepted for inclusion in Wayne State University Dissertations by an authorized administrator of DigitalCommons@WayneState. ESTROGEN SULFOTRANSFERASE (SULT1E1) EXPRESSION AND FUNCTION IN MCF10A-SERIES BREAST EPITHELIAL CELLS: ROLE AS A MODIFIER OF BREAST CARCINOGENESIS AND REGULATION BY PROLIFERATION STATE by JIAQI FU DISSERTATION Submitted to the Graduate School of Wayne State University, Detroit, Michigan in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY 2011 MAJOR: MOLECULAR AND CELLULAR TOXICOLOGY Approved by: Advisor Date DEDICATION I dedicate this work to my parents for all the sacrifices they have made on my behalf. To Luan, for his love and support during the preparation of this work. ii ACKNOWLEDGEMENTS I owe my deepest gratitude to Dr. Melissa Runge-Morris and Dr. Thomas A. Kocarek, for the encouragement and support throughout all these years of study. Their guidance helped me in all the time of research and writing of this thesis. -

Der Jasmonsäure-Metabolismus in Tomate (Solanum Lycopersicum
Literaturverzeichnis 6 Literaturverzeichnis 141 Literaturverzeichnis Afitlhile, M. M.; Fukushige, H.; Nishimura, M.; Hildebrand, D. F. (2005): A defect in glyoxysomal fatty acid ß-oxidation reduces jasmonic acid accumulation in Arabidopsis. Plant Physiology and Biochemistry 43: 603-609. Ahn, J. A.; Miller, D.; Winter, V. J.; Banfield, M. J.; Lee, J. H.; Yoo, S. Y.; Henz, S. R.; Brady, R. L.; Wei- gel, D. (2006): A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO Journal 25: 605–614. Allen, K. D.; Sussex, I. M. (1996): Falsiflora and anantha control early stages of floral meristem develop- ment in tomato (Lycopersicon esculentum Mill). Planta 200: 254–264. Ananvoranich, S.; Varin, L.; Gulick, P.; Ibrahim, R. K. (1994) Cloning and regulation of flavonol 3-sul- fotransferasein cell suspension cultures of Flaveria bidentis. Plant Physiol. 106: 485-491. Andersson, M. X.; Hamberg, M.; Kourtchenko, O.; Brunnstrom, A.; McPhail, K. L.; Gerwick, W. H.; Gobel, C., Feussner, I.; Ellerstrom, M. (2006): Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: formation of a novel oxo-phytodienoic acid-containing galactolipid, Arabidopside E. Journal of Biological Chemistry 281: 31528-31537. Atherton, J. G.; Harris, J. (1986): The tomato crop. London: Chapman and Hall. Barron, D.; Varin, L.; Ibrahim, R. K.; Harborne, J. B.; Williams, C. A. (1988): Sulphated flavonoids – an update. Phytochemistry 27: 2375-2395. Bäurle, I.; Dean, C. (2006): The timing of developmental transitions in plants. Cell 125: 655-664. Bell, P. R. (1992): Green plants: their origin and diversity. Cambridge: Cambridge University Press. Bergey, D. R.; Howe, G. A.; Ryan, C. -

Comparative Gene Expression of Intestinal Metabolizing Enzymes
BIOPHARMACEUTICS & DRUG DISPOSITION Biopharm. Drug Dispos. 30: 411–421 (2009) Published online 9 September 2009 in Wiley InterScience (www.interscience.wiley.com) DOI: 10.1002/bdd.675 Comparative Gene Expression of Intestinal Metabolizing Enzymes Ho-Chul Shina,Ã, Hye-Ryoung Kima, Hee-Jung Choa, Hee Yia, Soo-Min Choa, Dong-Goo Leea, A. M. Abd El-Atya, Jin-Suk Kima, Duxin Sunb and Gordon L . Amidonb aDepartment of Veterinary Pharmacology and Toxicology, Konkuk University, Seoul 143-701, Republic of Korea bDepartment of Pharmaceutical Sciences, College of Pharmacy, University of Michigan, Ann Arbor, MI 48109, USA ABSTRACT: The purpose of this study was to compare the expression profiles of drug- metabolizing enzymes in the intestine of mouse, rat and human. Total RNA was isolated from the duodenum and the mRNA expression was measured using Affymetrix GeneChip oligonucleotide arrays. Detected genes from the intestine of mouse, rat and human were ca. 60% of 22690 sequences, 40% of 8739 and 47% of 12559, respectively. Total genes of metabolizing enzymes subjected in this study were 95, 33 and 68 genes in mouse, rat and human, respectively. Of phase I enzymes, the mouse exhibited abundant gene expressions for Cyp3a25, Cyp4v3, Cyp2d26, followed by Cyp2b20, Cyp2c65 and Cyp4f14, whereas, the rat showed higher expression profiles of Cyp3a9, Cyp2b19, Cyp4f1, Cyp17a1, Cyp2d18, Cyp27a1 and Cyp4f6. However, the highly expressed P450 enzymes were CYP3A4, CYP3A5, CYP4F3, CYP2C18, CYP2C9, CYP2D6, CYP3A7, CYP11B1 and CYP2B6 in the human. For phase II enzymes, glucuronosyltransferase Ugt1a6, glutathione S-transferases Gstp1, Gstm3 and Gsta2, sulfotransferase Sult1b1 and acyltransferase Dgat1 were highly expressed in the mouse. -

Organellar Proteomics of the Golgi Apparatus and Golgi Derived COPI Vesicles
Organellar Proteomics of the Golgi Apparatus and Golgi Derived COPI Vesicles. Catherine Elaine Au Department of Anatomy and Cell Biology McGill University, Montreal January 2008 A thesis submitted to McGill University in partial fulfillment of the requirements of the degree of Doctor of Philosophy. ©Catherine Elaine Au 2008 Table of Contents Table of Contents .................................................................................................1 List of Figures .......................................................................................................7 List of Tables ........................................................................................................9 List of Abbreviations ...........................................................................................10 Abstract...............................................................................................................13 Resumé ..............................................................................................................14 Original Contributions .........................................................................................18 Acknowledgements.............................................................................................19 Chapter 1 -- Introduction.....................................................................................21 Chapter 2 -- Literature Review............................................................................24 2.1 The early secretory pathway. ....................................................... -

Structural and Biochemical Studies of Sulphotransferase 18 From
Structural and biochemical studies of sulphotransferase 18 from Arabidopsis thaliana explain its substrate specificity and reaction mechanism Hirschmann, Felix; Krause, Florian; Baruch, Petra; Chizhov, Igor; Mueller, Jonathan Wolf; Manstein, Dietmar J; Papenbrock, Jutta; Fedorov, Roman DOI: 10.1038/s41598-017-04539-2 License: Creative Commons: Attribution (CC BY) Document Version Publisher's PDF, also known as Version of record Citation for published version (Harvard): Hirschmann, F, Krause, F, Baruch, P, Chizhov, I, Mueller, JW, Manstein, DJ, Papenbrock, J & Fedorov, R 2017, 'Structural and biochemical studies of sulphotransferase 18 from Arabidopsis thaliana explain its substrate specificity and reaction mechanism', Scientific Reports, vol. 7, 4160. https://doi.org/10.1038/s41598-017-04539- 2 Link to publication on Research at Birmingham portal General rights Unless a licence is specified above, all rights (including copyright and moral rights) in this document are retained by the authors and/or the copyright holders. The express permission of the copyright holder must be obtained for any use of this material other than for purposes permitted by law. •Users may freely distribute the URL that is used to identify this publication. •Users may download and/or print one copy of the publication from the University of Birmingham research portal for the purpose of private study or non-commercial research. •User may use extracts from the document in line with the concept of ‘fair dealing’ under the Copyright, Designs and Patents Act 1988 (?) •Users may not further distribute the material nor use it for the purposes of commercial gain. Where a licence is displayed above, please note the terms and conditions of the licence govern your use of this document. -

Regulations of Expressions of Rat/Human Sulfotransferases (Sults) by Anti-Cancer Drug, Nolatrexed and Micronutrients
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.19.049007; this version posted April 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Regulations of expressions of rat/human sulfotransferases (SULTs) by anti-cancer drug, nolatrexed and micronutrients. Smarajit Maitia,b Sangita MaitiDuttac, Guangping Chend, aCell and Molecular Therapeutics Laboratory, Department of Biochemistry and Biotechnology, Oriental Institute of Science and Technology, Midnapore-721102, West Bengal, India. bFounder and Secretary, Agricure Biotech Research Society, Epidemiology and Human Health Division, Midnapore-721101, India. cDepartment of Biological Sciences, Midnapore City College, Midnapore, West Bengal, India. dVenture I OSU Laboratory, Oklahoma Technology & Research Park, 1110 S. Innovation Way, Stillwater, OK 74074, USA Correspondence: *Dr. Smarajit Maiti Professor and Head Post Graduate Department of Biochemistry and Biotechnology, Cell & Molecular Therapeutics Lab, OIST, Midnapore-721102, E. Mail: [email protected], Mobile: 9474504269 Running title: Nolatrexed induces Sulfotransferase expression Conflict of interests: None bioRxiv preprint doi: https://doi.org/10.1101/2020.04.19.049007; this version posted April 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Cancer is a disease related to cellular proliferative-state. Drastically increase in cell-cycle regulations augments cellular folate-pool and folate-metabolism.