Defining the Role of the NADH–Cytochrome-B5 Reductase 3 in the Mitochondrial Amidoxime Reducing Component Enzyme System
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

CYB5R3 Gene Cytochrome B5 Reductase 3
CYB5R3 gene cytochrome b5 reductase 3 Normal Function The CYB5R3 gene provides instruction for making an enzyme called cytochrome b5 reductase 3. This enzyme is involved in transferring negatively charged particles called electrons from one molecule to another. Two versions (isoforms) of this enzyme are produced from the CYB5R3 gene. The soluble isoform is present only in red blood cells, and the membrane-bound isoform is found in all other cell types. Normal red blood cells contain molecules of iron-containing hemoglobin, which deliver oxygen to the body's tissues. The iron in hemoglobin is ferrous (Fe2+), but it can spontaneously become ferric (Fe3+). Hemoglobin that contains ferric iron is called methemoglobin, and it cannot deliver oxygen. The soluble isoform of cytochrome b5 reductase 3 changes ferric iron back to ferrous iron so hemoglobin can function. Normally, red blood cells contain less than 2 percent methemoglobin. The membrane-bound isoform is embedded in the membranes of various cellular compartments and is widely used in the body. This isoform is necessary for many chemical reactions, including the breakdown and formation of fatty acids, the formation of cholesterol, and the breakdown of various molecules and drugs. Health Conditions Related to Genetic Changes Autosomal recessive congenital methemoglobinemia More than 65 mutations in the CYB5R3 gene have been found to cause autosomal recessive congenital methemoglobinemia types I and II. Most of these CYB5R3 gene mutations cause autosomal recessive congenital methemoglobinemia type I, which is characterized by a lack of oxygen in the body's tissues and bluish appearance of the skin, lips, and nails (cyanosis). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Mutations in SDHD Lead to Autosomal Recessive Encephalomyopathy And
Downloaded from http://jmg.bmj.com/ on December 5, 2017 - Published by group.bmj.com Genotype-phenotype correlations ORIGINAL ARTICLE Mutations in SDHD lead to autosomal recessive encephalomyopathy and isolated mitochondrial complex II deficiency Christopher Benjamin Jackson,1,2 Jean-Marc Nuoffer,3 Dagmar Hahn,3 Holger Prokisch,4,5 Birgit Haberberger,4 Matthias Gautschi,3,6 Annemarie Häberli,3 Sabina Gallati,1 André Schaller1 ▸ Additional material is ABSTRACT succinate to fumarate, is formed by the two larger published online only. To view Background Defects of the mitochondrial respiratory hydrophilic subunits, SDHA and SDHB, which also please visit the journal online (10.1136/jmedgenet-2013- chain complex II (succinate dehydrogenase (SDH) harbour the redox cofactors that participate in elec- 101932) complex) are extremely rare. Of the four nuclear encoded tron transfer to ubiquinone. The cofactor FAD is 1 proteins composing complex II, only mutations in the covalently bound to SDHA which provides the suc- Division of Human Genetics, fl fi Departments of Paediatrics and 70 kDa avoprotein (SDHA) and the recently identi ed cinate binding site, and SDHB possesses three Fe-S Clinical Research, University of complex II assembly factor (SDHAF1) have been found to centres which mediate the electron transfer to ubi- Bern, Bern, Switzerland be causative for mitochondrial respiratory chain diseases. quinone. The smaller hydrophobic SDHC and 2 Graduate School for Cellular Mutations in the other three subunits (SDHB, SDHC, SDHD subunits constitute the membrane anchor and Biomedical Sciences, SDHD) and the second assembly factor (SDHAF2) have and ubiquinone binding sites of CII and localise University of Bern, Bern, 2–4 Switzerland so far only been associated with hereditary CII to the inner mitochondrial membrane. -

Electron Transport Discovery Four Complexes Complex I: Nadhà Coqh2
BI/CH 422/622 OUTLINE: Pyruvate pyruvate dehydrogenase Krebs’ Cycle How did he figure it out? Overview 8 Steps Citrate Synthase Aconitase Isocitrate dehydrogenase Ketoglutarate dehydrogenase Succinyl-CoA synthetase Succinate dehydrogenase Fumarase Malate dehydrogenase Energetics Regulation Summary Oxidative Phosphorylation Energetics (–0.16 V needed for making ATP) Mitochondria Transport (2.4 kcal/mol needed to transport H+ out) Electron transport Discovery Four Complexes Complex I: NADHà CoQH2 Complex II: Succinateà CoQH2 2+ Complex III: CoQH2à Cytochrome C (Fe ) 2+ Complex IV: Cytochrome C (Fe ) à H2O Electron Transport à O2 Inhibitors of Electron Transport Big Drop! • Inhibitors all stop ET and ATP synthesis: very toxic! Spectral work Big Drop! NADH Cyto-a3 Cyto-c1 Big Drop! Cyto-b Cyto-c Cyto-a Fully reduced Flavin Cyto-c + rotenone + antimycin A 300 350 400 450 500 550 600 650 700 1 Electron Transport Electron-Transport Chain Complexes Contain a Series of Electron Carriers • Better techniques for isolating and handling mitochondria, and isolated various fractions of the inner mitochondrial membrane • Measure E°’ • They corresponded to these large drops, and they contained the redox compounds isolated previously. • When assayed for what reactions they could perform, they could perform certain redox reactions and not others. • When isolated, including isolating the individual redox compounds, and measuring the E°’ for each, it was clear that an electron chain was occurring; like a wire! • Lastly, when certain inhibitors were added, some of the redox reactions could be inhibited and others not. Site of the inhibition could be mapped. Electron Transport Electron-Transport Chain Complexes Contain a Series of Electron Carriers • Better techniques for isolating and handling mitochondria, and isolated various fractions of the inner mitochondrial membrane • Measure E°’ • They corresponded to these large drops, and they contained the redox compounds isolated previously. -
Supplementary Tables and Figures
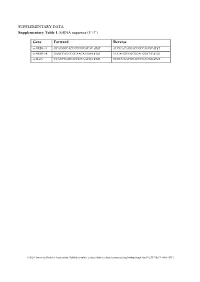
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC -
![And Detoxification (Benzola]Pyrene Quinones/Oxygen Radicals/NADPH-Cytochrome P-450 Reductase) PAUL L](https://docslib.b-cdn.net/cover/7447/and-detoxification-benzola-pyrene-quinones-oxygen-radicals-nadph-cytochrome-p-450-reductase-paul-l-837447.webp)
And Detoxification (Benzola]Pyrene Quinones/Oxygen Radicals/NADPH-Cytochrome P-450 Reductase) PAUL L
Proc. Nati. Acad. Sci. USA Vol. 81, pp. 1696-1700, March 1984 Biochemistry Mutagenicity of quinones: Pathways of metabolic activation and detoxification (benzola]pyrene quinones/oxygen radicals/NADPH-cytochrome P-450 reductase) PAUL L. CHESIS*, DAVID E. LEVIN*, MARTYN T. SMITHt, LARS ERNSTERt, AND BRUCE N. AMES* Departments of *Biochemistry and tBiomedical and Environmental Health Sciences, School of Public Health, University of California, Berkeley, CA 94720; and tDepartment of Biochemistry, Arrhenius Laboratory, University of Stockholm, S-10691 Stockholm, Sweden Contributed by Bruce N. Ames, December 12, 1983 ABSTRACT The mutagenicity of various quinones, a class nones might also be mutagenic, and we have tested this pos- of compounds widely distributed in nature, is demonstrated in sibility using the TA104 strain, which is sensitive to a wide the Salmonella TA104 tester strain. The metabolic pathways variety of oxidative mutagens (17). We have also attempted by which four quinones, menadione, benzo[alpyrene 3,6-qui- to characterize the pathways by which several different qui- none, 9,10-phenanthrenequinone, and danthron, caused mu- nones are metabolized and to study the potential mutagenic- tagenicity in this test system were investigated in detail as were ity of the metabolites and side products formed. We there- the detoxification pathways. The two-electron reduction of fore decided to investigate only those quinones that required these quinones by NAD(P)H-quinone oxidoreductase (DT-di- metabolic activation to exhibit mutagenicity. To limit the aphorase) was not mutagenic, whereas the one-electron reduc- scope of this project we also chose not to study quinones tion, catalyzed by NADPH-cytochrome P-450 reductase, was that possess reactive leaving groups. -

Cryoelectron Microscopy Structure and Mechanism of the Membrane-Associated Electron-Bifurcating Flavoprotein Fix/Etfabcx
Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX Xiang Fenga, Gerrit J. Schutb, Gina L. Lipscombb, Huilin Lia,1, and Michael W. W. Adamsb,1 aDepartment of Structural Biology, Van Andel Institute, Grand Rapids, MI 49503 and bDepartment of Biochemistry and Molecular Biology, University of Georgia, Athens, GA 30602 Edited by Michael A. Marletta, University of California, Berkeley, CA, and approved November 22, 2020 (received for review August 10, 2020) The electron-transferring flavoprotein-menaquinone oxidoreduc- FBEB enzymes can be divided into four independently tase ABCX (EtfABCX), also known as FixABCX for its role in evolved groups that are unrelated phylogenetically: 1) electron nitrogen-fixing organisms, is a member of a family of electron- transfer flavoproteins containing EtfAB; 2) [FeFe]-hydrogenases transferring flavoproteins that catalyze electron bifurcation. and formate dehydrogenases containing HydABC; 3) hetero- EtfABCX enables endergonic reduction of ferredoxin (E°′ ∼−450 disulfide reductases containing HdrA; and 4) transhydrogenases mV) using NADH (E°′ −320 mV) as the electron donor by coupling containing NfnAB. X-ray–based structures have been reported this reaction to the exergonic reduction of menaquinone (E°′ −80 for three of the four classes: for butyryl-CoA dehydrogenase mV). Here we report the 2.9 Å structure of EtfABCX, a membrane- (EtfAB-Bcd) (5) and caffeyl-CoA reductase [CarCDE; where associated flavin-based electron bifurcation (FBEB) complex, from CarDE corresponds to EtfAB (6)], for NfnAB (7), and for the a thermophilic bacterium. EtfABCX forms a superdimer with two MvhADG–HdrABC complex (8). No structural information is membrane-associated EtfCs at the dimer interface that contain available for a bifurcating HydABC-type enzyme. -

Discovery of Oxidative Enzymes for Food Engineering. Tyrosinase and Sulfhydryl Oxi- Dase
Dissertation VTT PUBLICATIONS 763 1,0 0,5 Activity 0,0 2 4 6 8 10 pH Greta Faccio Discovery of oxidative enzymes for food engineering Tyrosinase and sulfhydryl oxidase VTT PUBLICATIONS 763 Discovery of oxidative enzymes for food engineering Tyrosinase and sulfhydryl oxidase Greta Faccio Faculty of Biological and Environmental Sciences Department of Biosciences – Division of Genetics ACADEMIC DISSERTATION University of Helsinki Helsinki, Finland To be presented for public examination with the permission of the Faculty of Biological and Environmental Sciences of the University of Helsinki in Auditorium XII at the University of Helsinki, Main Building, Fabianinkatu 33, on the 31st of May 2011 at 12 o’clock noon. ISBN 978-951-38-7736-1 (soft back ed.) ISSN 1235-0621 (soft back ed.) ISBN 978-951-38-7737-8 (URL: http://www.vtt.fi/publications/index.jsp) ISSN 1455-0849 (URL: http://www.vtt.fi/publications/index.jsp) Copyright © VTT 2011 JULKAISIJA – UTGIVARE – PUBLISHER VTT, Vuorimiehentie 5, PL 1000, 02044 VTT puh. vaihde 020 722 111, faksi 020 722 4374 VTT, Bergsmansvägen 5, PB 1000, 02044 VTT tel. växel 020 722 111, fax 020 722 4374 VTT Technical Research Centre of Finland, Vuorimiehentie 5, P.O. Box 1000, FI-02044 VTT, Finland phone internat. +358 20 722 111, fax + 358 20 722 4374 Edita Prima Oy, Helsinki 2011 2 Greta Faccio. Discovery of oxidative enzymes for food engineering. Tyrosinase and sulfhydryl oxi- dase. Espoo 2011. VTT Publications 763. 101 p. + app. 67 p. Keywords genome mining, heterologous expression, Trichoderma reesei, Aspergillus oryzae, sulfhydryl oxidase, tyrosinase, catechol oxidase, wheat dough, ascorbic acid Abstract Enzymes offer many advantages in industrial processes, such as high specificity, mild treatment conditions and low energy requirements. -

Thioredoxin Reductase Two Modes of Catalysis Have Evolved
Eur. J. Biochem. 267, 6110±6117 (2000) q FEBS 2000 MINIREVIEW Thioredoxin reductase Two modes of catalysis have evolved Charles H. Williams Jr1,2, L. David Arscott1, Sylke MuÈ ller2,3, Brett W. Lennon4, Martha L. Ludwig2,4, Pan-Fen Wang2, Donna M. Veine1, Katja Becker5,* and R. Heiner Schirmer5 1Department of Veterans Affairs Medical Center, Ann Arbor, MI, USA; 2Department of Biological Chemistry, University of Michigan, Ann Arbor, MI, USA; 3Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany; 4Biophysics Research Division, University of Michigan, Ann Arbor, MI, USA, 5Biochemistry Center (BZH), Heidelberg University, Heidelberg, Germany Thioredoxin reductase (EC 1.6.4.5) is a widely distributed flavoprotein that catalyzes the NADPH-dependent reduction of thioredoxin. Thioredoxin plays several key roles in maintaining the redox environment of the cell. Like all members of the enzyme family that includes lipoamide dehydrogenase, glutathione reductase and mercuric reductase, thioredoxin reductase contains a redox active disulfide adjacent to the flavin ring. Evolution has produced two forms of thioredoxin reductase, a protein in prokaryotes, archaea and lower eukaryotes having a Mr of 35 000, and a protein in higher eukaryotes having a Mr of 55 000. Reducing equivalents are transferred from the apolar flavin binding site to the protein substrate by distinct mechanisms in the two forms of thioredoxin reductase. In the low Mr enzyme, interconversion between two conformations occurs twice in each catalytic cycle. After reduction of the disulfide by the flavin, the pyridine nucleotide domain must rotate with respect to the flavin domain in order to expose the nascent dithiol for reaction with thioredoxin; this motion repositions the pyridine ring adjacent to the flavin ring. -

Bioinformatics Analysis Based on Gene Expression Omnibus
ANTICANCER RESEARCH 39 : 1689-1698 (2019) doi:10.21873/anticanres.13274 Chemo-resistant Gastric Cancer Associated Gene Expression Signature: Bioinformatics Analysis Based on Gene Expression Omnibus JUN-BAO LIU 1* , TUNYU JIAN 2* , CHAO YUE 3, DAN CHEN 4, WEI CHEN 5, TING-TING BAO 6, HAI-XIA LIU 7, YUN CAO 8, WEI-BING LI 6, ZHIJIAN YANG 9, ROBERT M. HOFFMAN 9 and CHEN YU 6 1Traditional Chinese Medicine Department, People's Hospital of Henan Province, People's Hospital of Zhengzhou University, Zhengzhou, P.R. China; 2Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, P.R. China; 3Department of general surgery, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 4Research Center of Clinical Oncology, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 5Department of Head and Neck Surgery, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 6Department of Integrated TCM & Western Medicine, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 7Emergency Department, The Second Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, P.R. China; 8Master candidate of Oncology, Nanjing University of Chinese Medicine, Nanjing, P.R. China; 9AntiCancer, Inc., San Diego, CA, U.S.A. Abstract. Background/Aim: This study aimed to identify identified, including 13 up-regulated and 1,473 down-regulated biomarkers for predicting the prognosis of advanced gastric genes. -

Expression in Escherichia Coli of Cytochrome C Reductase Activity from a Maize NADH:Nitrate Reductase Complementary DNA'
Plant Physiol. (1992) 99, 693-699 Received for publication September 20, 1991 0032-0889/92/99/0693/07/$01 .00/0 Accepted December 11, 1991 Expression in Escherichia coli of Cytochrome c Reductase Activity from a Maize NADH:Nitrate Reductase Complementary DNA' Wilbur H. Campbell Phytotechnology Research Center and Department of Biological Sciences, Michigan Technological University, Houghton, Michigan 49931-1295 ABSTRACT the deduced amino acid sequences of NR to related mam- A cDNA clone was isolated from a maize (Zea mays L. cv malian protein sequences because of its similarity to the Mo- W64AxW183E) scutellum Xgtl 1 library using maize leaf pterin domain of sulfite oxidase, and the C-terminal region NADH:nitrate reductase Zmnrl cDNA clone as a hybridization of NR has sequence homology to NADH:Cyt b5 reductase, probe; it was designated Zmnr1S. ZmnrlS was shown to be an which is a FAD-containing enzyme (3, 22). In between the NADH:nitrate reductase clone by nucleotide sequencing and com- Mo-pterin and FAD domains, NR has similarity to mam- parison of its deduced amino acid sequence to Zmnrl. Zmnr1S, malian Cyt b5. This central Cyt b domain of NR is bridged to which is 1.8 kilobases in length and contains the code for both the other two domains on either side of it by highly variable the cytochrome b and flavin adenine dinucleotide domains of sequence regions that appear to be hinges (3). These hinge nitrate reductase, was cloned into the EcoRI site of the Esche- regions are probably exposed in native NR and susceptible to richia coli expression vector pET5b and expressed. -

Cytochrome C, Cytochrome B5 and Electron Transfer
Cytochrome c, cytochrome b5: electron transfer and new functions 1 Iron Protoporphyrin IX, heme b Heme a Prosthetic group Heme a is a form of heme found A flat and planar structure. In cytochromes a and a3. -. Itself Toxic O2 , H2O2 are formed. Insoluble 2 Covalent bonds Cytochrome c is a major player in membrane associated electron transport systems in bacteria and mitochondria. 3 Myoglobin, Hemoglobin His E7 Distal side Heme iron His F8 Proximal side Myoglobin has a strong affinity for oxygen Heme is located in the hydrophobic site. when it is in the lungs, and where the pressure is around 100 torr. When it reaches Heme Fe interacts with His. the tissues, where it's around 20 torr, the Coordination bond affinity for oxygen is still quite high. This makes myoglobin less efficient of an oxygen Propionates of heme interact with Arg. transporter than hemoglobin, which loses it's Ionic bond affinity for oxygen as the pressure goes down and releases the oxygen into the tissues. Heme is easily dissociated from the protein. Myoglobin's strong affinity for oxygen means that it keeps the oxygen binded to itself His (E7) is the oxygen binding site. instead of releasing it into the tissues. 4 Sickle cell anemia & malaria resistance In sickle cell hemoglobin (HbS) glutamic acid in position 6 (in beta chain) is mutated to valine. This change allows the deoxygenated form of the hemoglobin to stick to itself. 5 Figure 1. HO-1 Protects against Severe Malaria Using a mouse model for cerebral malaria, Ferreira et al. (2011) suggest a biochemical basis for the link between sickle cell disease and severe malaria.