Induced Pluripotent Stem Cells and Human Primary Immunodeficiencies
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-
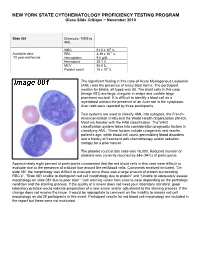
10 11 Cyto Slides 81-85
NEW YORK STATE CYTOHEMATOLOGY PROFICIENCY TESTING PROGRAM Glass Slide Critique ~ November 2010 Slide 081 Diagnosis: MDS to AML 9 WBC 51.0 x 10 /L 12 Available data: RBC 3.39 x 10 /L 72 year-old female Hemoglobin 9.6 g/dL Hematocrit 29.1 % MCV 86.0 fL Platelet count 16 x 109 /L The significant finding in this case of Acute Myelogenous Leukemia (AML) was the presence of many blast forms. The participant median for blasts, all types was 88. The blast cells in this case (Image 081) are large, irregular in shape and contain large prominent nucleoli. It is difficult to identify a blast cell as a myeloblast without the presence of an Auer rod in the cytoplasm. Auer rods were reported by three participants. Two systems are used to classify AML into subtypes, the French- American-British (FAB) and the World Health Organization (WHO). Most are familiar with the FAB classification. The WHO classification system takes into consideration prognostic factors in classifying AML. These factors include cytogenetic test results, patient’s age, white blood cell count, pre-existing blood disorders and a history of treatment with chemotherapy and/or radiation therapy for a prior cancer. The platelet count in this case was 16,000. Reduced number of platelets was correctly reported by 346 (94%) of participants. Approximately eight percent of participants commented that the red blood cells in this case were difficult to evaluate due to the presence of a bluish hue around the red blood cells. Comments received included, “On slide 081 the morphology was difficult to evaluate since there was a large amount of protein surrounding RBC’s”, “Slide 081 unable to distinguish red cell morphology due to protein” and “Unable to adequately assess morphology on slide 081 due to poor stain”. -
Blood Lab First Week
Blood Lab First Week This is a self propelled powerpoint study atlas of blood cells encountered in examination of the peripheral blood smear. It is a requirement of this course that you begin review of these slides with the first day of class in order to familiarize yourself with terms used in describing peripheral blood morphology. Laboratory final exam questions are derived directly from these slides. The content of these slides may duplicate slides (1-37) listed on the Hematopathology Website. You must familiarize yourself with both sets of slides in order to have the best learning opportunity. Before Beginning This Slide Review • Please read the Laboratory information PDF under Laboratory Resources file to learn about – Preparation of a blood smear – How to select an area of the blood smear for review of morphology and how to perform a white blood cell differential – Platelet estimation – RBC morphology descriptors • Anisocytosis • Poikilocytosis • Hypochromia • Polychromatophilia Normal blood smear. Red blood cells display normal orange pink cytoplasm with central pallor 1/3-1/2 the diameter of the red cell. There is mild variation in size (anisocytosis) but no real variation in shape (poikilocytosis). To the left is a lymphocyte. To the right is a typical neutrophil with the usual 3 segmentations of the nucleus. Med. Utah pathology Normal blood: thin area Ref 2 Normal peripheral blood smear. This field is good for exam of cell morphology, although there are a few minor areas of overlap of red cells. Note that most cells are well dispersed and the normal red blood cell central pallor is noted throughout the smear. -

ORIGINAL ARTICLE PML–Rara Initiates Leukemia by Conferring Properties of Self-Renewal to Committed Promyelocytic Progenitors
Leukemia (2009) 23, 1462–1471 & 2009 Macmillan Publishers Limited All rights reserved 0887-6924/09 $32.00 www.nature.com/leu ORIGINAL ARTICLE PML–RARa initiates leukemia by conferring properties of self-renewal to committed promyelocytic progenitors S Wojiski1,2, FC Guibal3, T Kindler1,2, BH Lee1,2, JL Jesneck4,5, A Fabian1,2, DG Tenen3,6 and DG Gilliland1,2,6 1Division of Hematology, Brigham and Women’s Hospital, Boston, MA, USA; 2Department of Genetics, Howard Hughes Medical Institute, Harvard Medical School, Boston, MA, USA; 3Department of Hematology/Oncology, Harvard Institutes of Medicine, Boston, MA, USA; 4Cancer Program, Broad Institute of Harvard and MIT, Cambridge, MA, USA; 5Department of Pediatric Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA and 6Harvard Stem Cell Institute, Harvard University, Boston, MA, USA Acute promyelocytic leukemia (APL) is characterized by the leukemia stem cells (LSCs) are rare cells contained within hyperproliferation of promyelocytes, progenitors that are the more primitive CD34 þ CD38À population, resembling committed to terminal differentiation into granulocytes, making it an ideal disease in which to study the transforming potential transformed hematopoietic stem cells (HSCs). LSC activity was of less primitive cell types. We utilized a murine model of APL in shown by these investigators in all AML subtypes tested, but which the PML–RARa oncogene is expressed from the were not shown in acute promyelocytic leukemia (APL) endogenous cathepsin G promoter to test the hypothesis that associated with the expression of the PML–RARa fusion as a leukemia stem cell (LSC) activity resides within the differen- consequence of an acquired t(15;17). -

Difference Between Promyelocyte and Myelocyte Key Difference
Difference Between Promyelocyte and Myelocyte www.differencebetween.com Key Difference - Promyelocyte vs Myelocyte Granulated blood cells include eosinophils, basophils, and neutrophils which participate in a variety of functions in the body. The precursor stem cells of these cells which arise from the hematopoietic stem cells are of the myeloid lineage. Myeloblasts are the precursor cells of granulated blood cells. Myeloblasts then mature into promyelocytes, myelocytes, metamyelocytes, bands, and segments to finally give rise to granulocytes in the peripheral blood tissue. The development process is known as Granulopoiesis. Promyelocyte is the second stage of Myeloblast development. Myelocyte is the third stage of Myeloblast development. The key difference between the promyelocyte and the myelocyte is the level of differentiation it exhibits. Promyelocytes do not show differentiation while myelocytes show differentiation. What is a Promyelocyte? Promyelocyte is the second stage of the Myeloblast development process. The promyelocyte is larger than the myeloblast. It has a diameter of 12-25µm and is the largest cell type in the myeloid series. It has a prominent nucleus, and the nucleus is placed slightly intended in the cytoplasm. Chromatin and nucleoli are prominent in this. Final structures of chromatin can be identified through microscopic observations. Towards the complete maturation of the promyelocyte, the chromatins appear as well-condensed structures. The condensed chromatin is placed along the nuclear membrane. The cytoplasm of the promyelocyte is granulated, and these granules are termed as the primary azurophilic granules. Since the promyelocyte is not differentiated, it is formed of a basophilic cytoplasm. The cell organelle organization is prominent in the promyelocyte stage of the blood cells. -

Advanced Blood Cell Identification
ADVANCED BLOOD CELL ID: LEUKOCYTES AND ERYTHROCYTES IN AN ACUTE LEUKEMIA Educational commentary is provided for participants enrolled in program #259- Advanced Blood Cell Identification. This virtual blood cell identification program includes case studies with more difficult challenges. To view the blood cell images in more detail, click on the sample identification numbers underlined in the paragraphs below. This will open a virtual image of the selected cell and the surrounding fields. If the image opens in the same window as the commentary, saving the commentary PDF and opening it outside your browser will allow you to switch between the commentary and the images more easily. Click on this link for the API ImageViewerTM Instructions. Learning Outcomes After completing this exercise, participants should be able to: • Discuss morphologic characteristics of normal peripheral blood leukocytes. • Describe morphologic features of immature granulocytes. • Identify morphologic abnormalities in erythrocyte shape and chromaticity/coloration. Case Study An 18 year old male was seen by his physician for bruising and severe nosebleeds. His CBC results are as follows: WBC=7.5 x 109/L, RBC=2.79 1012/L, Hgb=8.7 g/dL, Hct=24.6%, MCV=88 fL, MCH=31 pg, MCHC=35 g/dL, RDW=18.4%, Platelet=41 x 109/L, MPV=10.4. Educational Commentary The cells selected for identification and discussion in this exercise are from the peripheral blood smear of an 18 year old man diagnosed with acute promyelocytic leukemia (APL). APL is also referred to as acute myeloid leukemia, M3 (AML-M3). As with other acute leukemias, APL has a rapid onset. -

Henry Ford Hospital Clinicopathological Conference: Metamorphosis in Chronic Granulocytic Leukemia in a 60-Year-Old Man
Henry Ford Hospital Medical Journal Volume 28 Number 1 Article 7 3-1980 Henry Ford Hospital Clinicopathological Conference: Metamorphosis in chronic granulocytic leukemia in a 60-year-old man Follow this and additional works at: https://scholarlycommons.henryford.com/hfhmedjournal Part of the Life Sciences Commons, Medical Specialties Commons, and the Public Health Commons Recommended Citation (1980) "Henry Ford Hospital Clinicopathological Conference: Metamorphosis in chronic granulocytic leukemia in a 60-year-old man," Henry Ford Hospital Medical Journal : Vol. 28 : No. 1 , 37-46. Available at: https://scholarlycommons.henryford.com/hfhmedjournal/vol28/iss1/7 This Article is brought to you for free and open access by Henry Ford Health System Scholarly Commons. It has been accepted for inclusion in Henry Ford Hospital Medical Journal by an authorized editor of Henry Ford Health System Scholarly Commons. Henry Ford Hosp Med Journal Vol 28, No 1, 1980 Henry Ford Hospital Clinicopathological Conference Metamorphosis in chronic granulocytic leukemia in a 60-year-old man Participants: Protocol: Dr. Ellis J. Van Slyck, Department of Internal Medicine, Division of Hematology Discussant: Dr. Robert K. Nixon, Department of Internal Medicine, Second Medical Division Radiology: Dr. Mark C. Weingarden, Department of Diagnostic Radiology Pathology: Dr. John W. Rebuck, Department of Pathology, Division of Hematology Case Presentation As an outpatient, his myleran dosage was gradually tapered as the This 60-year-old white man came to Henry Ford Hospital in blood counts responded. On February 27, the white count was December 1975, complaining of increased fatigability over the 40,000 and the platelet count 400,000. On May 10, the white preceding year. -

Perforin-2 Is Essential for Intracellular Defense of Parenchymal Cells And
RESEARCH ARTICLE elifesciences.org Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria Ryan M McCormack1, Lesley R de Armas1, Motoaki Shiratsuchi1, Desiree G Fiorentino1, Melissa L Olsson1, Mathias G Lichtenheld1, Alejo Morales1, Kirill Lyapichev1, Louis E Gonzalez1, Natasa Strbo1, Neelima Sukumar2, Olivera Stojadinovic3, Gregory V Plano1, George P Munson1, Marjana Tomic- Canic3, Robert S Kirsner3, David G Russell2, Eckhard R Podack1* 1Department of Microbiology and Immunology, Miller School of Medicine, University of Miami, Miami, United States; 2Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, United States; 3Wound Healing and Regenerative Medicine Research Program, Department of Dermatology and Cutaneous Surgery, Miller School of Medicine, University of Miami, Miami, United States Abstract Perforin-2 (MPEG1) is a pore-forming, antibacterial protein with broad-spectrum activity. Perforin-2 is expressed constitutively in phagocytes and inducibly in parenchymal, tissue-forming cells. In vitro, Perforin-2 prevents the intracellular replication and proliferation of bacterial pathogens in these cells. Perforin-2 knockout mice are unable to control the systemic dissemination of methicillin-resistant Staphylococcus aureus (MRSA) or Salmonella typhimurium and perish shortly after epicutaneous or orogastric infection respectively. In contrast, Perforin-2-sufficient littermates clear the infection. Perforin-2 is a transmembrane protein of cytosolic vesicles -derived from multiple organelles- that translocate to and fuse with bacterium containing vesicles. Subsequently, Perforin-2 polymerizes and forms large clusters of 100 A˚ pores in the bacterial surface with Perforin-2 cleavage products present in bacteria. Perforin-2 is also required for the bactericidal activity of reactive oxygen and nitrogen species and hydrolytic enzymes. -

Bone Marrow Aspirate A Bone Marrow Film Should First Be Examined Macroscopically to Make Sure That Particles Or Fragments Are Present
Aspiration of the BM Satisfactory samples can usually be aspirated from the Sternum Anterior or posterior iliac spines Aspiration from only one site can give rise to misleading information; this is particularly true in aplastic anaemia as the marrow may be affected partially. There is little advantage in aspirating more than 0.3 ml of marrow fluid from a single site for morphological examination as this increases peripheral blood dilution. Bone marrow aspirate A bone marrow film should first be examined macroscopically to make sure that particles or fragments are present. Bone marrow aspirates which lack particles may be diluted with peripheral blood and may therefore be unrepresentative. An ideal bone marrow film with particles is shown. Even films without fragments are worth examining as useful information may be gained. However, assessment of cellularity and megakaryocyte numbers is unreliable and dilution with peripheral blood may lead to lymphocytes and neutrophils being over- represented in the differential count. Erythroid series Myeloid series Megakaryocytic series *Proerythroblast **Early erythroblast ***Intermediate erythroblast ****Late erythroblast Erythroid precursors Normal red cells are produced in the bone marrow from erythroid precursors or erythroblasts. The earliest morphologically recognisable red cell precursor is derived from an erythroid progenitor cell which in turn is derived from a multipotent haemopoietic progenitor cell. proerythroblast Normal proerythroblast [dark red arrow] in the bone marrow. This is a large cell with a round nucleus and a finely stippled chromatin pattern. Nucleoli are sometimes apparent. The cytoplasm is moderately to strongly basophilic. There may be a paler staining area of cytoplasm surrounding the nucleus. Normal erythroblasts in the BM . -

62754323.Pdf
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by University of Missouri: MOspace Hemopoiesis The process of red cell production is called erythropoiesis, during which the erythrocyte undergoes progressive changes that involve the cytoplasm and nucleus. The cell progressively becomes smaller, and the cytoplasm stains increasingly acidophilic as it accumulates hemoglobin and loses organelles. The nucleus shrinks and becomes more heterochromatic and condensed until ultimately it is lost from the cell. Although it is possible to describe various "stages" in the developmental sequence, the process of erythropoiesis does not occur in stepwise fashion. The process is a continuous one in which, at several points, the cells show distinctive, recognizable features. Unfortunately, the nomenclature of the red cell precursors is confused by the multiplicity of names given by various investigators to stages in the maturational series. The terms used here are commonly used, but alternative nomenclature is provided. The proerythroblast (pronormoblast, rubriblast) is the earliest recognizable precursor of the red cell line and is derived from the pluripotent stem cell through a series of restricted stem cells. The proerythroblast is relatively large, with a diameter of 15 to 20 µm. The nongranular, basophilic cytoplasm frequently stains unevenly, showing patches that are relatively poorly stained, especially in a zone close to the nucleus. The cytoplasmic basophilia is an important point in the identification of this early form of red cell. Synthesis of hemoglobin has begun, but its presence is obscured by the basophilia of the cytoplasm. The nucleus occupies almost three-fourths of the cell body, and its chromatin is finely and uniformly granular or stippled in appearance. -

Platelet Morphology Received January 20, 2020; Accepted March 28, 2020 Introduction
J Lab Med 2020; 44(5): 231–239 Review Christoph Robier* Platelet morphology https://doi.org/10.1515/labmed-2020-0007 Received January 20, 2020; accepted March 28, 2020 Introduction Abstract The examination of a peripheral blood (PB) smear should be requested in every case of unexplained thrombocytope- Background: The examination of a peripheral blood smear nia or thrombocytosis. First, the morphological analysis is mandatory in case of unexplained thrombocytopenia should include the estimation of the number of platelets or thrombocytosis. First, the number of platelets should in order to confirm the platelet count determined by the be estimated in order to confirm the platelet count deter- haematology analyser, and to rule out causes of spuriously mined by the haematology analyser, and to rule out causes low platelet counts due to platelet aggregates, platelet sat- of spuriously low or elevated platelet counts. Second, the ellitism or platelet phagocytosis [1–4]. The identification size and morphological features of the platelets, which of pseudothrombocytopenia (PTP) due to anticoagulant- may provide information on the underlying cause of the induced platelet aggregation is crucial, because a misin- low or enhanced platelet count, have to be assessed. terpretation as a “true” thrombocytopenia may lead to Content: This review summarizes the physiological and serious diagnostic and therapeutic consequences such as pathological features of platelet size and morphology, bone marrow biopsy, initiation of corticosteroid therapy, circulating megakaryocytes, micromegakaryocytes and platelet transfusion or even splenectomy [2]. PTP can be megakaryoblasts, and provides an overview of current caused by various anticoagulants such as ethylenedi- guidelines on the reporting of platelet morphology. -

Acute Promyelocytic Leukemia
Acute promyelocytic leukemia Description Acute promyelocytic leukemia is a form of acute myeloid leukemia, a cancer of the blood-forming tissue (bone marrow). In normal bone marrow, hematopoietic stem cells produce red blood cells (erythrocytes) that carry oxygen, white blood cells (leukocytes) that protect the body from infection, and platelets (thrombocytes) that are involved in blood clotting. In acute promyelocytic leukemia, immature white blood cells called promyelocytes accumulate in the bone marrow. The overgrowth of promyelocytes leads to a shortage of normal white and red blood cells and platelets in the body, which causes many of the signs and symptoms of the condition. People with acute promyelocytic leukemia are especially susceptible to developing bruises, small red dots under the skin (petechiae), nosebleeds, bleeding from the gums, blood in the urine (hematuria), or excessive menstrual bleeding. The abnormal bleeding and bruising occur in part because of the low number of platelets in the blood ( thrombocytopenia) and also because the cancerous cells release substances that cause excessive bleeding. The low number of red blood cells (anemia) can cause people with acute promyelocytic leukemia to have pale skin (pallor) or excessive tiredness (fatigue). In addition, affected individuals may heal slowly from injuries or have frequent infections due to the loss of normal white blood cells that fight infection. Furthermore, the leukemic cells can spread to the bones and joints, which may cause pain in those areas. Other general signs and symptoms may occur as well, such as fever, loss of appetite, and weight loss. Acute promyelocytic leukemia is most often diagnosed around age 40, although it can be diagnosed at any age. -

This Lab Has an Associated Homework Assignment That Is Posted on Blackboard
LABORATORY 13 - BLOOD CELL DEVELOPMENT This lab has an associated homework assignment that is posted on Blackboard. If you have not done this assignment already, it would make this lab more efficient if you did so. OBJECTIVES: LIGHT MICROSCOPY: Recognize the different stages of erythrocyte development: proerythroblast, basophilic erythroblast, polychromatophilic erythroblast, orthochromatophilic erythroblast and reticulocyte. Recognize the different stages of granulocyte development: myeloblast, promyelocyte, neutrophilic and eosinophilic myelocytes, neutrophilic and eosinophilic metamyelocytes, and neutrophilic band and neutrophilic segmented cells. Recognize and understand the significance of the morphological changes that occur during development of these cells. ELECTRON MICROSCOPY: See Section F ASSIGNMENT FOR TODAY'S LABORATORY GLASS SLIDES SL 177 Peripheral blood smear from a patient with chronic myelogenous leukemia (CML), to be used for identifying early stages of granulocyte development - use tables and key at the end of this section. SL 68 Normal bone marrow smear - Stages of development of erythrocytes and granulocytes use tables and key at the end of this section. SL 22 Peripheral blood smear of baby's blood, reticulocytes SL 39 Adult bone marrow SL 41 Adult bone marrow ELECTRON MICROGRAPHS See Section F HISTOLOGY IMAGE REVIEW - available on computers in HSL Chapter 6. Blood and Myeloid Tissue Frames: 326-344 SUPPLEMENTARY ELECTRON MICROGRAPHS Rhodin, J. A.G., An Atlas of Histology Copies of this text are on reserve in the HSL. Megakaryocytes pp. 77 and 78 13 - 1 BLOOD CELL DEVELOPMENT Review the developmental stages of each cell type from the earliest form to mature cells by studying Figure 13-5 in Junqueira. Follow each cell type from the earliest recognizable stage to the mature cell type.