Myelin and Nodal Plasticity Modulate Action Potential Conduction in The
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Vocabulario De Morfoloxía, Anatomía E Citoloxía Veterinaria
Vocabulario de Morfoloxía, anatomía e citoloxía veterinaria (galego-español-inglés) Servizo de Normalización Lingüística Universidade de Santiago de Compostela COLECCIÓN VOCABULARIOS TEMÁTICOS N.º 4 SERVIZO DE NORMALIZACIÓN LINGÜÍSTICA Vocabulario de Morfoloxía, anatomía e citoloxía veterinaria (galego-español-inglés) 2008 UNIVERSIDADE DE SANTIAGO DE COMPOSTELA VOCABULARIO de morfoloxía, anatomía e citoloxía veterinaria : (galego-español- inglés) / coordinador Xusto A. Rodríguez Río, Servizo de Normalización Lingüística ; autores Matilde Lombardero Fernández ... [et al.]. – Santiago de Compostela : Universidade de Santiago de Compostela, Servizo de Publicacións e Intercambio Científico, 2008. – 369 p. ; 21 cm. – (Vocabularios temáticos ; 4). - D.L. C 2458-2008. – ISBN 978-84-9887-018-3 1.Medicina �������������������������������������������������������������������������veterinaria-Diccionarios�������������������������������������������������. 2.Galego (Lingua)-Glosarios, vocabularios, etc. políglotas. I.Lombardero Fernández, Matilde. II.Rodríguez Rio, Xusto A. coord. III. Universidade de Santiago de Compostela. Servizo de Normalización Lingüística, coord. IV.Universidade de Santiago de Compostela. Servizo de Publicacións e Intercambio Científico, ed. V.Serie. 591.4(038)=699=60=20 Coordinador Xusto A. Rodríguez Río (Área de Terminoloxía. Servizo de Normalización Lingüística. Universidade de Santiago de Compostela) Autoras/res Matilde Lombardero Fernández (doutora en Veterinaria e profesora do Departamento de Anatomía e Produción Animal. -

Oligodendrocytes in Development, Myelin Generation and Beyond
cells Review Oligodendrocytes in Development, Myelin Generation and Beyond Sarah Kuhn y, Laura Gritti y, Daniel Crooks and Yvonne Dombrowski * Wellcome-Wolfson Institute for Experimental Medicine, Queen’s University Belfast, Belfast BT9 7BL, UK; [email protected] (S.K.); [email protected] (L.G.); [email protected] (D.C.) * Correspondence: [email protected]; Tel.: +0044-28-9097-6127 These authors contributed equally. y Received: 15 October 2019; Accepted: 7 November 2019; Published: 12 November 2019 Abstract: Oligodendrocytes are the myelinating cells of the central nervous system (CNS) that are generated from oligodendrocyte progenitor cells (OPC). OPC are distributed throughout the CNS and represent a pool of migratory and proliferative adult progenitor cells that can differentiate into oligodendrocytes. The central function of oligodendrocytes is to generate myelin, which is an extended membrane from the cell that wraps tightly around axons. Due to this energy consuming process and the associated high metabolic turnover oligodendrocytes are vulnerable to cytotoxic and excitotoxic factors. Oligodendrocyte pathology is therefore evident in a range of disorders including multiple sclerosis, schizophrenia and Alzheimer’s disease. Deceased oligodendrocytes can be replenished from the adult OPC pool and lost myelin can be regenerated during remyelination, which can prevent axonal degeneration and can restore function. Cell population studies have recently identified novel immunomodulatory functions of oligodendrocytes, the implications of which, e.g., for diseases with primary oligodendrocyte pathology, are not yet clear. Here, we review the journey of oligodendrocytes from the embryonic stage to their role in homeostasis and their fate in disease. We will also discuss the most common models used to study oligodendrocytes and describe newly discovered functions of oligodendrocytes. -

Neuregulin 1–Erbb2 Signaling Is Required for the Establishment of Radial Glia and Their Transformation Into Astrocytes in Cerebral Cortex
Neuregulin 1–erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex Ralf S. Schmid*, Barbara McGrath*, Bridget E. Berechid†, Becky Boyles*, Mark Marchionni‡, Nenad Sˇ estan†, and Eva S. Anton*§ *University of North Carolina Neuroscience Center and Department of Cell and Molecular Physiology, University of North Carolina School of Medicine, Chapel Hill, NC 27599; †Department of Neurobiology, Yale University School of Medicine, New Haven, CT 06510; and ‡CeNes Pharamceuticals, Inc., Norwood, MA 02062 Communicated by Pasko Rakic, Yale University School of Medicine, New Haven, CT, January 27, 2003 (received for review December 12, 2002) Radial glial cells and astrocytes function to support the construction mine whether NRG-1-mediated signaling is involved in radial and maintenance, respectively, of the cerebral cortex. However, the glial cell development and differentiation in the cerebral cortex. mechanisms that determine how radial glial cells are established, We show that NRG-1 signaling, involving erbB2, may act in maintained, and transformed into astrocytes in the cerebral cortex are concert with Notch signaling to exert a critical influence in the not well understood. Here, we show that neuregulin-1 (NRG-1) exerts establishment, maintenance, and appropriate transformation of a critical role in the establishment of radial glial cells. Radial glial cell radial glial cells in cerebral cortex. generation is significantly impaired in NRG mutants, and this defect can be rescued by exogenous NRG-1. Down-regulation of expression Materials and Methods and activity of erbB2, a member of the NRG-1 receptor complex, leads Clonal Analysis to Study NRG’s Role in the Initial Establishment of to the transformation of radial glial cells into astrocytes. -

Pre-Oligodendrocytes from Adult Human CNS
The Journal of Neuroscience, April 1992, 12(4): 1538-l 547 Pre-Oligodendrocytes from Adult Human CNS Regina C. Armstrong,lJ Henry H. Dorn, l,b Conrad V. Kufta,* Emily Friedman,3 and Monique E. Dubois-Dalcq’ ‘Laboratory of Viral and Molecular Pathogenesis, and %urgical Neurology Branch, National Institute of Neurological Disorders and Stroke, Bethesda, Maryland 20892 and 3Department of Neurosurgery, University of Pennsylvania, Philadelphia, Pennsylvania 19104-3246 CNS remyelination and functional recovery often occur after Rapid and efficient neurotransmission is dependent upon the experimental demyelination in adult rodents. This has been electrical insulating capacity of the myelin sheath around axons attributed to the ability of mature oligodendrocytes and/or (reviewed in Ritchie, 1984a,b). Nerve conduction is impaired their precursor cells to divide and regenerate in response after loss of the myelin sheath and results in severe neurological to signals in demyelinating lesions. To determine whether dysfunction in human demyelinating diseases such as multiple oligodendrocyte precursor cells exist in the adult human sclerosis (MS). Remyelination can occur in the CNS of MS CNS, we have cultured white matter from patients under- patients but appears to be limited (Perier and Gregoire, 1965; going partial temporal lobe resection for intractable epilep- Prineas et al., 1984). Studies of acute MS cases have revealed sy. These cultures contained a population of process-bear- that recent demyelinating lesions can exhibit remyelination that ing cells that expressed antigens recognized by the 04 appears to correlate with the generation of new oligodendrocytes monoclonal antibody, but these cells did not express galac- (Prineas et al., 1984; Raine et al., 1988). -

A System for Studying Mechanisms of Neuromuscular Junction Development and Maintenance Valérie Vilmont1,‡, Bruno Cadot1, Gilles Ouanounou2 and Edgar R
© 2016. Published by The Company of Biologists Ltd | Development (2016) 143, 2464-2477 doi:10.1242/dev.130278 TECHNIQUES AND RESOURCES RESEARCH ARTICLE A system for studying mechanisms of neuromuscular junction development and maintenance Valérie Vilmont1,‡, Bruno Cadot1, Gilles Ouanounou2 and Edgar R. Gomes1,3,*,‡ ABSTRACT different animal models and cell lines (Chen et al., 2014; Corti et al., The neuromuscular junction (NMJ), a cellular synapse between a 2012; Lenzi et al., 2015) with the hope of recapitulating some motor neuron and a skeletal muscle fiber, enables the translation of features of neuromuscular diseases and understanding the triggers chemical cues into physical activity. The development of this special of one of their common hallmarks: the disruption of the structure has been subject to numerous investigations, but its neuromuscular junction (NMJ). The NMJ is one of the most complexity renders in vivo studies particularly difficult to perform. studied synapses. It is formed of three key elements: the lower motor In vitro modeling of the neuromuscular junction represents a powerful neuron (the pre-synaptic compartment), the skeletal muscle (the tool to delineate fully the fine tuning of events that lead to subcellular post-synaptic compartment) and the Schwann cell (Sanes and specialization at the pre-synaptic and post-synaptic sites. Here, we Lichtman, 1999). The NMJ is formed in a step-wise manner describe a novel heterologous co-culture in vitro method using rat following a series of cues involving these three cellular components spinal cord explants with dorsal root ganglia and murine primary and its role is basically to ensure the skeletal muscle functionality. -
Thenerveimpulse05.Pdf
The nerve impulse. INTRODUCTION Axons are responsible for the transmission of information between different points of the nervous system and their function is analogous to the wires that connect different points in an electric circuit. However, this analogy cannot be pushed very far. In an electrical circuit the wire maintains both ends at the same electrical potential when it is a perfect conductor or it allows the passage of an electron current when it has electrical resistance. As we will see in these lectures, the axon, as it is part of a cell, separates its internal medium from the external medium with the plasma membrane and the signal conducted along the axon is a transient potential difference1 that appears across this membrane. This potential difference, or membrane potential, is the result of ionic gradients due to ionic concentration differences across the membrane and it is modified by ionic flow that produces ionic currents perpendicular to the membrane. These ionic currents give rise in turn to longitudinal currents closing local ionic current circuits that allow the regeneration of the membrane potential changes in a different region of the axon. This process is a true propagation instead of the conduction phenomenon occurring in wires. To understand this propagation we will study the electrical properties of axons, which include a description of the electrical properties of the membrane and how this membrane works in the cylindrical geometry of the axon. Much of our understanding of the ionic mechanisms responsible for the initiation and propagation of the action potential (AP) comes from studies on the squid giant axon by A. -

Spatial Extent of Neurons: Dendrites and Axons
Spatial extent of neurons: dendrites and axons Morphological diversity of neurons Cortical pyramidal cell Purkinje cell Retinal ganglion cells Motoneurons Dendrites: spiny vs non-spiny Recording and simulating dendrites Axons - myelinated vs unmyelinated Axons - myelinated vs unmyelinated • Myelinated axons: – Long-range axonal projections (motoneurons, long-range cortico-cortical connections in white matter, etc) – Saltatory conduction; – Fast propagation (10s of m/s) • Unmyelinated axons: – Most local axonal projections – Continuous conduction – Slower propagation (a few m/s) Recording from axons Recording from axons Recording from axons - where is the spike generated? Modeling neuronal processes as electrical cables • Axial current flowing along a neuronal cable due to voltage gradient: V (x + ∆x; t) − V (x; t) = −Ilong(x; t)RL ∆x = −I (x; t) r long πa2 L where – RL: total resistance of a cable of length ∆x and radius a; – rL: specific intracellular resistivity • ∆x ! 0: πa2 @V Ilong(x; t) = − (x; t) rL @x The cable equation • Current balance in a cylinder of width ∆x and radius a • Axial currents leaving/flowing into the cylinder πa2 @V @V Ilong(x + ∆x; t) − Ilong(x; t) = − (x + ∆x; t) − (x; t) rL @x @x • Ionic current(s) flowing into/out of the cell 2πa∆xIion(x; t) • Capacitive current @V I (x; t) = 2πa∆xc cap M @t • Kirschoff law Ilong(x + ∆x; t) − Ilong(x; t) + 2πa∆xIion(x; t) + Icap(x; t) = 0 • In the ∆x ! 0 limit: @V a @2V cM = 2 − Iion @t 2rL @x Compartmental appoach Compartmental approach Modeling passive dendrites: Cable equation -

Clustering of Na+ Channels and Node of Ranvier Formation in Remyelinating Axons
The Journal of Neuroscience, January 1995, 15(l): 492503 Clustering of Na+ Channels and Node of Ranvier Formation in Remyelinating Axons Sanja Dugandgija-NovakoviC,’ Adam G. Koszowski,2 S. Rock Levinson,2 and Peter Shragerl ‘Department of Physiology, University of Rochester Medical Center, Rochester, New York 14642 and 2Department of Physiology, University of Colorado Health Sciences Center, Denver, Colorado 80262 Polyclonal antibodies were raised against a well conserved nodal regions(Black et al., 1990). The density of Na+ channels, region of the vertebrate Na+ channel and were affinity pu- in particular, is about 25 times higher at nodesof Ranvier than rified for use in immunocytochemistry. Focal demyelination at internodal sites (Shrager, 1989). There has been vigorous of rat sciatic axons was initiated by an intraneural injection debate over the mechanism of Na+ channel clustering during of lysolecithin and Na+ channel clustering was followed at myelination, particularly with respect to the role of Schwann several stages of myelin removal and repair. At 1 week post- cells, and studies have included both developing nerve and injection axons contained long, fully demyelinated regions. pathological conditions (Ellisman, 1979; Rosenbluth, 1979; Ro- Na+ channel clusters appeared only at heminodes forming senbluth and Blakemore, 1984; Le Beau et al., 1987; England the borders of these zones, and at widely spaced isolated et al., 1990, 1991; Joe and Angelides, 1992, 1993).There remain sites that may represent former nodes of Ranvier. Over the many interesting questions, particularly regarding remodeling next few days proliferating Schwann cells adhered to axons that occurs following myelin disruption. When axons are de- and began to extend processes. -

Structure and Function Relationship in Nerve Cells & Membrane Potential
Structure and Function Relationship in Nerve Cells & Membrane Potential Asist. Prof. Aslı AYKAÇ NEU Faculty of Medicine Biophysics Nervous System Cells • Glia – Not specialized for information transfer – Support neurons • Neurons (Nerve Cells) – Receive, process, and transmit information Neurons • Neuron Doctrine – The neuron is the functional unit of the nervous system • Specialized cell type – have very diverse in structure and function Neuron: Structure/Function • designed to receive, process, and transmit information – Dendrites: receive information from other neurons – Soma: “cell body,” contains necessary cellular machinery, signals integrated prior to axon hillock – Axon: transmits information to other cells (neurons, muscles, glands) • Information travels in one direction – Dendrite → soma → axon How do neurons work? • Function – Receive, process, and transmit information – Conduct unidirectional information transfer • Signals – Chemical – Electrical Membrane Potential Because of motion of positive and negative ions in the body, electric current generated by living tissues. What are these electrical signals? • receptor potentials • synaptic potentials • action potentials Why are these electrical signals important? These signals are all produced by temporary changes in the current flow into and out of cell that drives the electrical potential across the cell membrane away from its resting value. The resting membrane potential The electrical membrane potential across the membrane in the absence of signaling activity. Two type of ions channels in the membrane – Non gated channels: always open, important in maintaining the resting membrane potential – Gated channels: open/close (when the membrane is at rest, most gated channels are closed) Learning Objectives • How transient electrical signals are generated • Discuss how the nongated ion channels establish the resting potential. -

Regulation of Myelin Structure and Conduction Velocity by Perinodal Astrocytes
Correction NEUROSCIENCE Correction for “Regulation of myelin structure and conduc- tion velocity by perinodal astrocytes,” by Dipankar J. Dutta, Dong Ho Woo, Philip R. Lee, Sinisa Pajevic, Olena Bukalo, William C. Huffman, Hiroaki Wake, Peter J. Basser, Shahriar SheikhBahaei, Vanja Lazarevic, Jeffrey C. Smith, and R. Douglas Fields, which was first published October 29, 2018; 10.1073/ pnas.1811013115 (Proc. Natl. Acad. Sci. U.S.A. 115,11832–11837). The authors note that the following statement should be added to the Acknowledgments: “We acknowledge Dr. Hae Ung Lee for preliminary experiments that informed the ultimate experimental approach.” Published under the PNAS license. Published online June 10, 2019. www.pnas.org/cgi/doi/10.1073/pnas.1908361116 12574 | PNAS | June 18, 2019 | vol. 116 | no. 25 www.pnas.org Downloaded by guest on October 2, 2021 Regulation of myelin structure and conduction velocity by perinodal astrocytes Dipankar J. Duttaa,b, Dong Ho Wooa, Philip R. Leea, Sinisa Pajevicc, Olena Bukaloa, William C. Huffmana, Hiroaki Wakea, Peter J. Basserd, Shahriar SheikhBahaeie, Vanja Lazarevicf, Jeffrey C. Smithe, and R. Douglas Fieldsa,1 aSection on Nervous System Development and Plasticity, The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892; bThe Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., Bethesda, MD 20817; cMathematical and Statistical Computing Laboratory, Office of Intramural Research, Center for Information -

View Software
TLR4-activated microglia have divergent effects on oligodendrocyte lineage cells DISSERTATION Presented in partial fulfillment of the requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University Evan Zachary Goldstein, BA Neuroscience Graduate Program The Ohio State University 2016 Dissertation Committee: Dr. Dana McTigue, Advisor Dr. Phillip Popovich Dr. Jonathan Godbout Dr. Courtney DeVries Copyright by Evan Zachary Goldstein 2016 i Abstract Myelin accelerates action potential conduction velocity and provides essential metabolic support for axons. Unfortunately, myelin and myelinating cells are often vulnerable to injury or disease, resulting in myelin damage, which in turn can lead to axon dysfunction, overt pathology and neurological impairment. Inflammation is a common component of CNS trauma and disease, and therefore an active inflammatory response is often considered deleterious to myelin health. While inflammation can certainly damage myelin, inflammatory processes also benefit oligodendrocyte (OL) lineage progression and myelin repair. Consistent with the divergent nature of inflammation, intraspinal toll-like receptor 4 (TLR4) activation, an innate immune pathway, kills OL lineage cells, but also initiates oligodendrogenesis. Soluble factors produced by TLR4-activated microglia can reproduce these effects in vitro, however the exact factors are unknown. To determine what microglial factors might contribute to TLR4-induced OL loss and oligodendrogenesis, mRNA of factors known to affect OL lineage cells was quantified in TLR4-activated microglia and spinal cords (chapter 2). Results indicate that TLR4-activated microglia transcribe numerous factors that induce OL loss, OL progenitor cell (OPC) proliferation and OPC differentiation. However, some factors upregulated after intraspinal TLR4 activation were not ii upregulated by microglia, suggesting that other cell types contribute to transcriptional changes in vivo. -
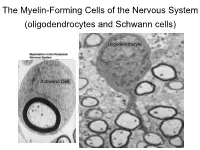
The Myelin-Forming Cells of the Nervous System (Oligodendrocytes and Schwann Cells)
The Myelin-Forming Cells of the Nervous System (oligodendrocytes and Schwann cells) Oligodendrocyte Schwann Cell Oligodendrocyte function Saltatory (jumping) nerve conduction Oligodendroglia PMD PMD Saltatory (jumping) nerve conduction Investigating the Myelinogenic Potential of Individual Oligodendrocytes In Vivo Sparse Labeling of Oligodendrocytes CNPase-GFP Variegated expression under the MBP-enhancer Cerebral Cortex Corpus Callosum Cerebral Cortex Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Ant Commissure Corpus Callosum Cerebral Cortex Caudate Putamen Piriform Cortex Corpus Callosum Ant Commissure Characterization of Oligodendrocyte Morphology Cerebral Cortex Corpus Callosum Caudate Putamen Cerebellum Brain Stem Spinal Cord Oligodendrocytes in disease: Cerebral Palsy ! CP major cause of chronic neurological morbidity and mortality in children ! CP incidence now about 3/1000 live births compared to 1/1000 in 1980 when we started intervening for ELBW ! Of all ELBW {gestation 6 mo, Wt. 0.5kg} , 10-15% develop CP ! Prematurely born children prone to white matter injury {WMI}, principle reason for the increase in incidence of CP ! ! 12 Cerebral Palsy Spectrum of white matter injury ! ! Macro Cystic Micro Cystic Gliotic Khwaja and Volpe 2009 13 Rationale for Repair/Remyelination in Multiple Sclerosis Oligodendrocyte specification oligodendrocytes specified from the pMN after MNs - a ventral source of oligodendrocytes