An Update on the Metabolic Roles of Carbonic Anhydrases in a Model
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Association Analyses of Known Genetic Variants with Gene
ASSOCIATION ANALYSES OF KNOWN GENETIC VARIANTS WITH GENE EXPRESSION IN BRAIN by Viktoriya Strumba A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Bioinformatics) in The University of Michigan 2009 Doctoral Committee: Professor Margit Burmeister, Chair Professor Huda Akil Professor Brian D. Athey Assistant Professor Zhaohui S. Qin Research Statistician Thomas Blackwell To Sam and Valentina Dmitriy and Elizabeth ii ACKNOWLEDGEMENTS I would like to thank my advisor Professor Margit Burmeister, who tirelessly guided me though seemingly impassable corridors of graduate work. Throughout my thesis writing period she provided sound advice, encouragement and inspiration. Leading by example, her enthusiasm and dedication have been instrumental in my path to becoming a better scientist. I also would like to thank my co-advisor Tom Blackwell. His careful prodding always kept me on my toes and looking for answers, which taught me the depth of careful statistical analysis. His diligence and dedication have been irreplaceable in most difficult of projects. I also would like to thank my other committee members: Huda Akil, Brian Athey and Steve Qin as well as David States. You did not make it easy for me, but I thank you for believing and not giving up. Huda’s eloquence in every subject matter she explained have been particularly inspiring, while both Huda’s and Brian’s valuable advice made the completion of this dissertation possible. I would also like to thank all the members of the Burmeister lab, both past and present: Sandra Villafuerte, Kristine Ito, Cindy Schoen, Karen Majczenko, Ellen Schmidt, Randi Burns, Gang Su, Nan Xiang and Ana Progovac. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

High-Throughput Screening Studies of Inhibition of Human Carbonic Anhydrase II and Bacterial Flagella Antimicrobial Activity
Western Michigan University ScholarWorks at WMU Dissertations Graduate College 5-2010 High-Throughput Screening Studies of Inhibition of Human Carbonic Anhydrase II and Bacterial Flagella Antimicrobial Activity Albert A. Barrese III Western Michigan University Follow this and additional works at: https://scholarworks.wmich.edu/dissertations Part of the Biochemistry, Biophysics, and Structural Biology Commons, and the Biology Commons Recommended Citation Barrese, Albert A. III, "High-Throughput Screening Studies of Inhibition of Human Carbonic Anhydrase II and Bacterial Flagella Antimicrobial Activity" (2010). Dissertations. 500. https://scholarworks.wmich.edu/dissertations/500 This Dissertation-Open Access is brought to you for free and open access by the Graduate College at ScholarWorks at WMU. It has been accepted for inclusion in Dissertations by an authorized administrator of ScholarWorks at WMU. For more information, please contact [email protected]. HIGH-THROUGHPUT SCREENING STUDIES OF INHIBITION OF HUMAN CARBONIC ANHYDRASE II AND BACTERIAL FLAGELLA ANTIMICROBIAL ACTIVITY by Albert A. Barrese III A Dissertation Submitted to the Faculty of The Graduate College in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Department of Biological Sciences Advisor: Brian C. Tripp, Ph.D. Western Michigan University Kalamazoo, Michigan May 2010 UMI Number: 3410393 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. UMT Dissertation Publishing UMI 3410393 Copyright 2010 by ProQuest LLC. -

Quantigene Flowrna Probe Sets Currently Available
QuantiGene FlowRNA Probe Sets Currently Available Accession No. Species Symbol Gene Name Catalog No. NM_003452 Human ZNF189 zinc finger protein 189 VA1-10009 NM_000057 Human BLM Bloom syndrome VA1-10010 NM_005269 Human GLI glioma-associated oncogene homolog (zinc finger protein) VA1-10011 NM_002614 Human PDZK1 PDZ domain containing 1 VA1-10015 NM_003225 Human TFF1 Trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) VA1-10016 NM_002276 Human KRT19 keratin 19 VA1-10022 NM_002659 Human PLAUR plasminogen activator, urokinase receptor VA1-10025 NM_017669 Human ERCC6L excision repair cross-complementing rodent repair deficiency, complementation group 6-like VA1-10029 NM_017699 Human SIDT1 SID1 transmembrane family, member 1 VA1-10032 NM_000077 Human CDKN2A cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) VA1-10040 NM_003150 Human STAT3 signal transducer and activator of transcripton 3 (acute-phase response factor) VA1-10046 NM_004707 Human ATG12 ATG12 autophagy related 12 homolog (S. cerevisiae) VA1-10047 NM_000737 Human CGB chorionic gonadotropin, beta polypeptide VA1-10048 NM_001017420 Human ESCO2 establishment of cohesion 1 homolog 2 (S. cerevisiae) VA1-10050 NM_197978 Human HEMGN hemogen VA1-10051 NM_001738 Human CA1 Carbonic anhydrase I VA1-10052 NM_000184 Human HBG2 Hemoglobin, gamma G VA1-10053 NM_005330 Human HBE1 Hemoglobin, epsilon 1 VA1-10054 NR_003367 Human PVT1 Pvt1 oncogene homolog (mouse) VA1-10061 NM_000454 Human SOD1 Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) -

An Update on the Metabolic Roles of Carbonic Anhydrases in the Model Alga Chlamydomonas Reinhardtii
H OH metabolites OH Review An Update on the Metabolic Roles of Carbonic Anhydrases in the Model Alga Chlamydomonas reinhardtii Ashok Aspatwar 1,* ID , Susanna Haapanen 1 and Seppo Parkkila 1,2 1 Faculty of Medicine and Life Sciences, University of Tampere, FI-33014 Tampere, Finland; [email protected].fi (S.H.); [email protected].fi (S.P.) 2 Fimlab, Ltd., and Tampere University Hospital, FI-33520 Tampere, Finland * Correspondence: [email protected].fi; Tel.: +358-46-596-2117 Received: 11 January 2018; Accepted: 10 March 2018; Published: 13 March 2018 Abstract: Carbonic anhydrases (CAs) are metalloenzymes that are omnipresent in nature. − + CAs catalyze the basic reaction of the reversible hydration of CO2 to HCO3 and H in all living organisms. Photosynthetic organisms contain six evolutionarily different classes of CAs, which are namely: α-CAs, β-CAs, γ-CAs, δ-CAs, ζ-CAs, and θ-CAs. Many of the photosynthetic organisms contain multiple isoforms of each CA family. The model alga Chlamydomonas reinhardtii contains 15 CAs belonging to three different CA gene families. Of these 15 CAs, three belong to the α-CA gene family; nine belong to the β-CA gene family; and three belong to the γ-CA gene family. The multiple copies of the CAs in each gene family may be due to gene duplications within the particular CA gene family. The CAs of Chlamydomonas reinhardtii are localized in different subcellular compartments of this unicellular alga. The presence of a large number of CAs and their diverse subcellular localization within a single cell suggests the importance of these enzymes in the metabolic and biochemical roles they perform in this unicellular alga. -

Table S1. Detailed Clinical Features of Individuals with IDDCA and LADCI Syndromes
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Med Genet Table S1. Detailed clinical features of individuals with IDDCA and LADCI syndromes Lodder E., De Nittis P., Koopman C. et al., 2016 Family A Family B Family C Family D Family E Family F Individual 1 2 3 4 5 6 7 8 9 Gender, Age (years) F, 22 F, 20 F, 6 F, 11 M, 9 F, 12 F, 13 M, 8 M, 23 c.249G>A,r.249_250 c.249G>A,r.249_250 Nucleotide change c.249+1G>T/ c.249+3G>T/ c.249+3G>T/ c.906C>G/ c.242C>T/ c.242C>T/ c.242C>T/ ins249+1_249+25/ ins249+1_249+25/ (NM_006578.3) c.249+1G>T c.249+3G>T c.249+3G>T c.906C>G c.242C>T c.242C>T c.242C>T c.994C>T c.994C>T Amino acid change p.Asp84Valfs*52/ p.Asp84Valfs*52/ p.Asp84Leufs*31/ p.Asp84Valfs*31/ p.Asp84Valfs*31/ p.Tyr302*/ p.(Ser81Leu)/ p.(Ser81Leu)/ p.(Ser81Leu)/ (NP_006569.1) p.(Arg332*) p.(Arg332*) p.Asp84Leufs*31 p.Asp84Valfs*31 p.Asp84Valfs*31 p.Tyr302* p.(Ser81Leu) p.(Ser81Leu) p.(Ser81Leu) 3580 g (50th 2751 g (15 th Birth weight NA NA NA 2845 g (15th NA NA NA percentile) percentile) percentile) Ethnicity Italy Italy Jordan Puerto Rico Puerto Rico India Morocco Morocco Brazil Consanguinity − − + + + − − − + Altered speech + + NR + + + + + NA development - Verbal NA NA nonverbal unremarkable unremarkable NA NA NA NA understanding - Lexical production NA NA nonverbal delayed delayed nonverbal delayed delayed NA Intellectual + + + + + + mild mild mild disability (ID) Epilepsy + + + - -

Dissertation Kollipara.Pdf
Characterizing protein processing in the Endoplasmic Reticulum using quantitative proteomics: the pathogenesis of the Marinesco-Sjörgren Syndrome Zur Erlangung des akademischen Grades eines Dr. rer. nat von der Fakultät Bio- und Chemieingenieurwesen der Technischen Universität Dortmund genehmigte Dissertation vorgelegt von MSc. Laxmikanth Kollipara aus Hyderabad, India Tag der mündlichen Prüfung: 20.12.2016 1. Gutachter: Prof. Dr. Albert Sickmann 2. Gutachter: Prof. Dr. Oliver Kayser Dortmund 2016 1. Prüfer: Prof. Dr. Markus Nett Abstract Abstract In this work, characterization of Marinesco-Sjögren Syndrome (MSS) was performed for the first time on the proteome level using mass spectrometry (MS)-based quantitative proteomics strategies. MSS is a neuromuscular and neurodegenerative disorder and it is caused due to the mutational inactivation of SIL1 protein, which results in malfunctioning of protein folding machinery mediated by the chaperone BiP that can lead to the ER stress-induced cell death via apoptotic signaling. The major goals were (i) to understand the rescue mechanisms in SIL1-deficient non-vulnerable tissues from human and (ii) to verify the cellular perturbations caused due to the loss of functional SIL1 in woozy mouse (i.e. mouse model of MSS). To achieve these aims, samples derived from five different MSS cases and two different tissues from woozy along with their respective healthy controls were studied. For this, comparative LC-MS proteomics approaches such as chemical labeling (i.e. iTRAQ) and label- free quantification (precursor ion intensity based and NSAF) were employed. During which, sample preparation workflows were optimized that enabled to process clinical samples related to MSS that included primary cell lines and mammalian tissues for the subsequent quantitative LC-MS analyses. -

S1 of S77 Supplementary Materials: Discovery of a New Class of Cathepsin Kinhibitors in Rhizoma Drynariaeas Potential Candidates for the Treatment of Osteoporosis
Int. J. Mol. Sci.2016, 17, 2116; doi:10.3390/ijms17122116 S1 of S77 Supplementary Materials: Discovery of a New Class of Cathepsin KInhibitors in Rhizoma Drynariaeas Potential Candidates for the Treatment of Osteoporosis Zuo-Cheng Qiu, Xiao-Li Dong, Yi Dai, Gao-Keng Xiao, Xin-Luan Wang, Ka-Chun Wong, Man-Sau Wong and Xin-Sheng Yao Table S1. Compounds identified from Drynariae rhizome (DR). No. Compound Name Chemical Structure 1 Naringin 5,7,3′,5′-Tetrahydroxy-flavanone 2 7-O-neohesperidoside 3 Narigenin-7-O-β-D-glucoside 5,7,3′,5′-Tetrahydroxy-flavanone 4 7-O-β-D-glucopyranoside 5 Naringenin 6 5,7,3′,5′-Tetrahydroxyflavanone 7 Kushennol F 8 Sophoraflavanone G 9 Kurarinone Int. J. Mol. Sci.2016, 17, 2116; doi:10.3390/ijms17122116 S2 of S77 Table S1. Cont. No. Compound Name Chemical Structure 10 Leachianone A 11 Luteolin-7-O-neohesperidoside 12 Luteolin-5-O-neohesperidoside 13 Kaempferol-7-O-α-L-arabinofuranoside 14 8-Prenylapigenin 15 Apigenine 16 Kaempferol-3-O-α-L-rhamnopyranoside OH HO O 17 Astragalin O OH OH O O OH OH OH 18 3-O-β-D-Glucopyranoside-7-O-α-L-arabinofuranoside OH HO O HO O O 19 5,7-Dihydroxychromone-7-O-β-D-glucopyranoside OH OH O Int. J. Mol. Sci.2016, 17, 2116; doi:10.3390/ijms17122116 S3 of S77 Table S1. Cont. No. Compound Name Chemical Structure 20 5,7-Dihydroxychromone-7-O-neohesperidoside Kaempferol 21 3-O-β-D-glucopyranoside-7-O-β-D-glucopyranoside 22 Xanthohumol OH HO O 23 Epicatechin OH OH OH 24 (E)-4-O-β-D-Glucopyranosyl caffeic acid 25 β-D-Glucopyranosyl sinapoic acid 26 4-O-β-D-Glucopyranosyl ferulic acid 27 Trans-caffeic acid 28 4-O-β-D-Glucopyranosyl coumaric acid 29 Dihydrocaffeic acid methyl ester 30 Dihydrocaffeic acid 31 3,4-Dihydroxyl benzoic acid 32 4-O-D-Glucosyl vanillic acid Int. -

Sex Differences in Circulating Proteins in Heart Failure with Preserved
Sex differences in circulating proteins in heart failure with preserved ejection fraction Susan Stienen, Joao Pedro Ferreira, Masatake Kobayashi, Gregoire Preud’homme, Daniela Dobre, Jean-Loup Machu, Kévin Duarte, Emmanuel Bresso, Marie-Dominique Devignes, Natalia López Andrés, et al. To cite this version: Susan Stienen, Joao Pedro Ferreira, Masatake Kobayashi, Gregoire Preud’homme, Daniela Dobre, et al.. Sex differences in circulating proteins in heart failure with preserved ejection fraction. Biologyof Sex Differences, BioMed Central, 2020, 11 (1), pp.47. 10.1186/s13293-020-00322-7. hal-02928742 HAL Id: hal-02928742 https://hal.univ-lorraine.fr/hal-02928742 Submitted on 16 Oct 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License 1 Sex-differences in circulating proteins in heart failure with preserved ejection fraction 2 Susan Stienen, MD, PhD1; João Pedro Ferreira, MD, PhD1,2; Masatake Kobayashi, MD1 ; Gregoire 3 Preud’homme MSc1; Daniela Dobre1,3, Jean-Loup Machu, MSc1; Kevin Duarte PhD1 ; Emmanuel Bresso, 4 PhD4; Marie-Dominique Devignes, PhD4; Natalia López Andrés, PhD5; Nicolas Girerd, MD, PhD1; Svend 5 Aakhus6,7; Giuseppe Ambrosio8 ; Hans-Peter Brunner-La Rocca, MD, PhD9 ; Ricardo Fontes-Carvalho10; 6 Alan G. -
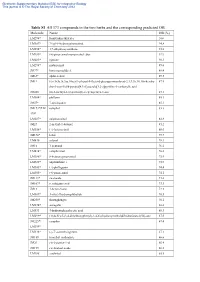
Table S1 All 373 Compounds in the Two Herbs and the Corresponding
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Table S1 All 373 compounds in the two herbs and the corresponding predicted OB Molecule Name OB (%) LM294* forsythidmethylester 100 LM387* 7'-epi-8-hydroxypinoresinol 94.8 LM394* 1,7-dihydroxyxanthone 93.8 LM289* (+)-pinoresinol monomethyl ether 92.9 LM285* egenine 90.3 LM298* matairesinol 89.6 JM77* loniceracetalide A 89.4 JM63* alpha-cedrol 89.3 JM1* (-)-(3r,8s,9r,9as,10as)-9-ethenyl-8-(beta-d-glucopyranosyloxy)-2,3,9,9a,10,10a-hexahy 87.5 dro-5-oxo-5h,8h-pyrano[4,3-d]oxazolo[3,2-a]pyridine-3-carboxylic acid JM200 (z)-3-methyl-2-(2-pentenyl)-2-cyclopenten-1-one 87.1 LM304* phillyrin 85.1 JM57* 7-epi-loganin 85.1 JM173*/LM camphol 83.3 398* LM287* epipinoresinol 82.8 JM27 2-methyl-1-butanol 81.2 LM356* (+)-lariciresinol 80.0 JM170* ledol 79.7 LM430 safynol 78.3 JM16 1-pentanol 76.2 LM414* campherenol 76.0 LM386* 8-hydroxypinoresinol 75.9 LM428* onjixanthone i 75.9 LM345* (+)-phillygenin 74.4 LM305* (+)-pinoresinol 74.1 JM113* sweroside 73.6 JM107* secologanic acid 73.3 JM13 1-hexen-3-one 72.3 LM309* 3-ethyl-7hydroxyphthalide 70.5 JM250* shuangkangsu 70.1 LM274* astragalin 68.6 LM331 4-hydroxyphenylacetic acid 68.3 LM299* (3r,4s,5r)-5-(3,4-dimethoxyphenyl)-3,4-bis(hydroxymethyl)dihydrofuran-2(3h)-one 67.5 JM225*/ camphor 67.4 LM399* LM338* (-)-7’-o-methylegenine 67.1 JM189 trimethyl isothiazole 66.6 JM29 cis-2-penten-1-ol 66.4 JM199 cis-linalool oxide 66.2 LM288 erythritol 65.8 Electronic Supplementary Material (ESI) for -

Network Pharmacology-Based Analysis of the Pharmacological Mechanisms of Aloperine on Cardiovascular Disease
Hindawi Evidence-Based Complementary and Alternative Medicine Volume 2020, Article ID 5180716, 8 pages https://doi.org/10.1155/2020/5180716 Research Article Network Pharmacology-Based Analysis of the Pharmacological Mechanisms of Aloperine on Cardiovascular Disease Bingwu Huang,1 Juncheng Xiong,1 Xuyong Zhao,2 Yihan Zheng,3 and Ning Zhu 2 1Department of Anesthesiology, e Wenzhou ird Clinical Institute Affiliated to Wenzhou Medical University, Wenzhou People’s Hospital, No. 299 Guan Road, Wenzhou 325000, Zhejiang Province, China 2Department of Cardiology, e Wenzhou ird Clinical Institute Affiliated to Wenzhou Medical University, Wenzhou People’s Hospital, No. 299 Guan Road, Wenzhou 325000, Zhejiang Province, China 3Eye Hospital, School of Ophthalmology & Optometry, Wenzhou Medical University, State Key Laboratory of Ophthalmology, Optometry and Visual Science, No. 270 Xueyuan West Road, Wenzhou 325000, Zhejiang Province, China Correspondence should be addressed to Ning Zhu; [email protected] Received 27 March 2020; Revised 25 May 2020; Accepted 18 June 2020; Published 14 July 2020 Academic Editor: Giuseppe Caminiti Copyright © 2020 Bingwu Huang et al. -is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Aloperine is an active component of Sophora alopecuroides Linn, which has been extensively applied for the treatment of cardiovascular disease (CVD). However, our current understanding of the molecular mechanisms supporting the effects of aloperine on CVD remains unclear. Methods. Systematic network pharmacology was conducted to provide testable hypotheses about pharmacological mechanisms of the protective effects of aloperine against CVD. Detailed structure was obtained from Traditional Chinese Medicines Integrated Database (TCMID). -

Cation-Coupled Bicarbonate Transporters
Cation-Coupled Bicarbonate Transporters Christian Aalkjaer, Aarhus University Ebbe Boedtkjer, Aarhus University Inyeong Choi, Emory University Soojung Lee, Emory University Journal Title: Comprehensive Physiology Volume: Volume 4, Number 4 Publisher: American Physiological Society | 2014-10-01, Pages 1605-1637 Type of Work: Article | Post-print: After Peer Review Publisher DOI: 10.1002/cphy.c130005 Permanent URL: https://pid.emory.edu/ark:/25593/rmnmf Final published version: http://dx.doi.org/10.1002/cphy.c130005 Copyright information: © 2013 American Physiological Society. All rights reserved. Accessed October 1, 2021 10:22 PM EDT HHS Public Access Author manuscript Author Manuscript Author ManuscriptCompr Physiol Author Manuscript. Author manuscript; Author Manuscript available in PMC 2016 February 26. Published in final edited form as: Compr Physiol. 2014 October ; 4(4): 1605–1637. Cation-Coupled Bicarbonate Transporters Christian Aalkjaer1,*, Ebbe Boedtkjer1, Inyeong Choi2, and Soojung Lee2 1Department of Biomedicine, and the Water and Salt Research Center, Aarhus University, Aarhus, Denmark 2Department of Physiology, Emory University School of Medicine, Atlanta, USA Abstract − Cation-coupled HCO3 transport was initially identified in the mid-1970s when pioneering studies − + showed that acid extrusion from cells is stimulated by CO2/HCO3 and associated with Na and Cl− movement. The first Na+-coupled bicarbonate transporter (NCBT) was expression-cloned in the late 1990s. There are currently five mammalian NCBTs in the SLC4-family: the electrogenic Na,HCO3-cotransporters NBCe1 and NBCe2 (SLC4A4 and SLC4A5 gene products); the + electroneutral Na,HCO3-cotransporter NBCn1 (SLC4A7 gene product); the Na -driven Cl,HCO3- exchanger NDCBE (SLC4A8 gene product); and NBCn2/NCBE (SLC4A10 gene product), which + has been characterized as an electroneutral Na,HCO3-cotransporter or a Na -driven Cl,HCO3- exchanger.