PDF (Dataset S01)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

List & Label Preview File
L I N A R I S I n f o r m a t i o n B I O L O G I S C H E P R O D U K T E PRIMÄRANTIKÖRPER-Veterinär erkennen: Maus, verschiedene Labels alphabetisch geordnet Beschreibung Format Klon Wirt Isotyp Anwendung Menge ME Kat.Nr. Description Format Clone Host Isotype Application Quantity Cat.No. Mouse Anti-Cardiolipin Ig's -ve control for ELISA 1 ml ADI-5502 IgG Mouse 0,5 mg ADI-AMPT11-M-500 Anti-Cardiolipin Ig's +ve control for ELISA polyclonal Mouse 1 ml ADI-5503 CD27 PE (Armenian Hamster IgG1) Hamster 50 tests ADI-MCD027-PE Monoclonal Anti-VSV-G-Cy conjugate for Mouse 0,1 mg ADI-VSV11-Cy Immunofluorescence Interleukin-4 IgG Mouse 0,1 mg ADI-AB-10710 Anti-Myelin Oligodendrocyte Glycoprotein IgG Mouse 0,1 mg ADI-AB-19910 CD160 mAb, PE, , (mouse IgG2bk) Mouse 50 tests ADI-MCD160-PE CD81, Purified (mouse IgG1) Mouse 0,1 mg ADI-MCD081-UL Interleukin-2 receptor IgG Rat 0,1 mg ADI-AB-10310 Interleukin-2 IgG Rat 0,1 mg ADI-AB-10510 Interleukin-4 IgG Rat 0,1 mg ADI-AB-10810 Interleukin-10 IgG Rat 0,1 mg ADI-AB-11310 Interleukin-12p40 IgG Rat 0,1 mg ADI-AB-11410 CTLA-4 IgG Rat 0,1 mg ADI-AB-11610 CD80 IgG Rat 0,1 mg ADI-AB-13310 CD11a IgG Rat 0,1 mg ADI-AB-13510 CD11b-FITC IgG Rat 0,1 mg ADI-AB-13610 B220 IgG Rat 0,1 mg ADI-AB-13910 CD90 Thy-1.1 IgG Rat 0,1 mg ADI-AB-14010 CD90 Thy-1.2 IgG Rat 0,1 mg ADI-AB-14110 CD90 Thy-1 IgG Rat 0,1 mg ADI-AB-14210 CD4 IgG Rat 0,1 mg ADI-AB-16510 IFN-gamma IgG Rat 0,1 mg ADI-AB-16610 CD3 IgG Rat 0,1 mg ADI-AB-17510 Interleukin-12p75 IgG Rat 0,1 mg ADI-AB-20910 CD8b , PE-Cy5 (Clone CT-CD8b) (rat IgG2a) Rat 50 tests -

Monoacylglycerol As a Metabolic Coupling Factor in Glucose-Stimulated Insulin Secretion
Université de Montréal Monoacylglycerol as a metabolic coupling factor in glucose-stimulated insulin secretion par Shangang Zhao Département de Biochimie Faculté de Médecine Mémoire présentée à la Faculté des Etudes Supérieures en vue de l’obtention du grade de maître ès sciences en Biochimie Décembre 2010 © Shangang Zhao, 2010 Université de Montréal Faculté des études supérieures Ce mémoire intitulée : Monoacylglycerol as a metabolic coupling factor in glucose-stimulated insulin secretion Présenté par : Shangang Zhao a été évaluée par un jury composé des personnes suivantes: Dr Tony Antakly, président-rapporteur Dr Marc Prentki, directeur de recherche Dr Ashok K. Srivastava, membre du jury i Résumé Les cellules beta pancréatiques sécrètent l’insuline lors d’une augmentation post-prandiale du glucose dans le sang. Ce processus essentiel est contrôlé par des facteurs physiologiques, nutritionnels et pathologiques. D’autres sources d’énergie, comme les acides aminés (leucine et glutamine) ou les acides gras potentialisent la sécrétion d’insuline. Une sécrétion d’insuline insuffisante au besoin du corps déclanche le diabète. Le rôle que joue l’augmentation du calcium intracellulaire et les canaux K+/ATP dans la sécrétion d’insuline est bien connu. Bien que le mécanisme exact de la potentialisation de la sécrétion d’insuline par les lipides est inconnu, le cycle Glycérolipides/Acides gras (GL/FFA) et son segment lipolytique ont été reconnu comme un composant essentiel de la potentialisation lipidique de la sécrétion d’insuline. Le diacylglycérol, provenant de la lipolyse, a été proposé comme un signal lipidique important d’amplification. Cependant, l’hydrolyse des triglycérides et des diacylglycérides a été démontrée essentielle pour la sécrétion d’insuline stimulée par le glucose, en suggérant un rôle du monoacylglycérol (MAG) dans ce processus. -

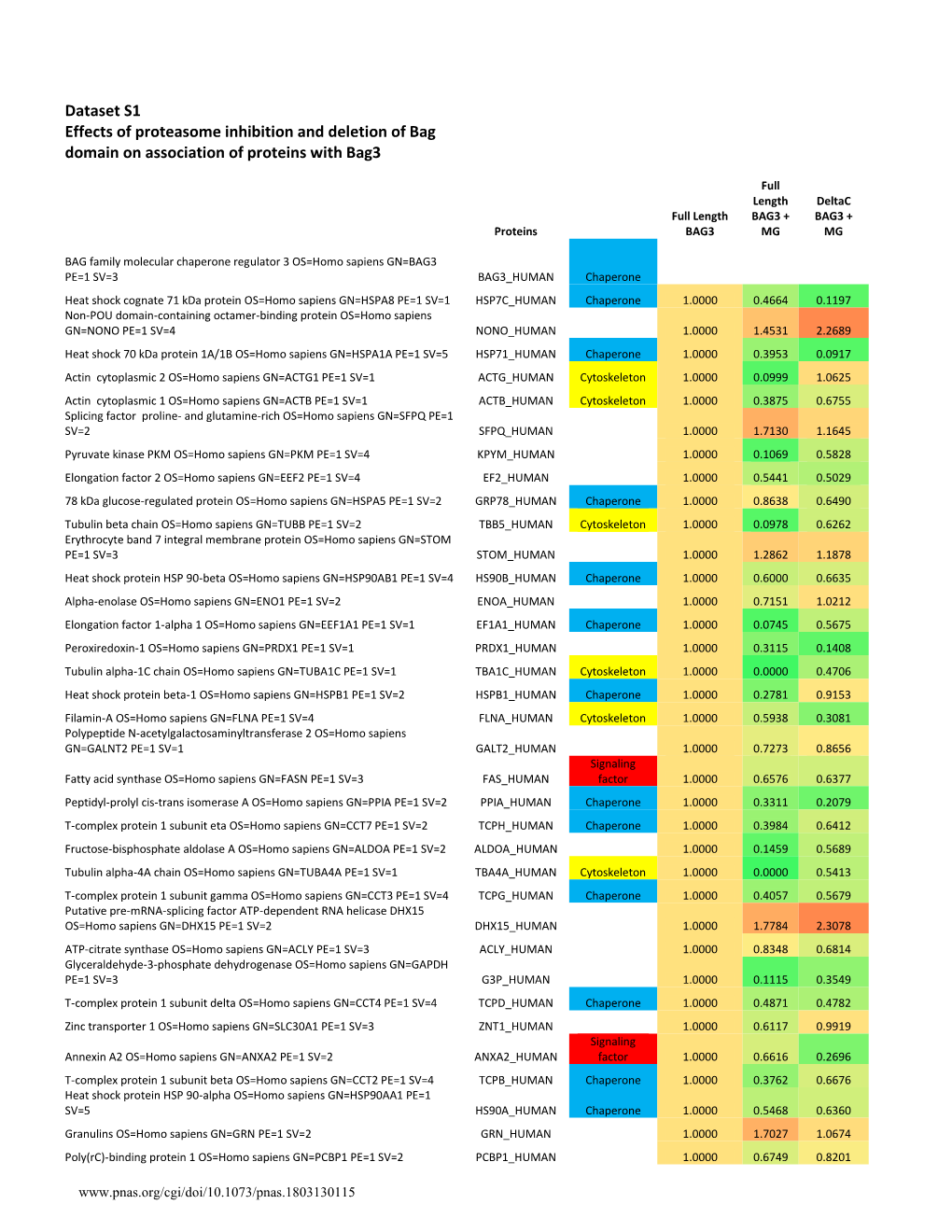
Supplemental Data
Article TCF7L2 is a master regulator of insulin production and processing ZHOU, Yuedan, et al. Abstract Genome-wide association studies have revealed >60 loci associated with type 2 diabetes (T2D), but the underlying causal variants and functional mechanisms remain largely elusive. Although variants in TCF7L2 confer the strongest risk of T2D among common variants by presumed effects on islet function, the molecular mechanisms are not yet well understood. Using RNA-sequencing, we have identified a TCF7L2-regulated transcriptional network responsible for its effect on insulin secretion in rodent and human pancreatic islets. ISL1 is a primary target of TCF7L2 and regulates proinsulin production and processing via MAFA, PDX1, NKX6.1, PCSK1, PCSK2 and SLC30A8, thereby providing evidence for a coordinated regulation of insulin production and processing. The risk T-allele of rs7903146 was associated with increased TCF7L2 expression, and decreased insulin content and secretion. Using gene expression profiles of 66 human pancreatic islets donors', we also show that the identified TCF7L2-ISL1 transcriptional network is regulated in a genotype-dependent manner. Taken together, these results demonstrate that not only synthesis of [...] Reference ZHOU, Yuedan, et al. TCF7L2 is a master regulator of insulin production and processing. Human Molecular Genetics, 2014, vol. 23, no. 24, p. 6419-6431 DOI : 10.1093/hmg/ddu359 PMID : 25015099 Available at: http://archive-ouverte.unige.ch/unige:45177 Disclaimer: layout of this document may differ from the published -

ANALYSIS of GENE PATHWAYS INVOLVED in DCIS PROGRESSION in RESPONSE to ACIDIC EXTRACELLULAR Ph Neha Aggarwal1, Jennifer Rothberg2, Robert J
ANALYSIS OF GENE PATHWAYS INVOLVED IN DCIS PROGRESSION IN RESPONSE TO ACIDIC EXTRACELLULAR pH Neha Aggarwal1, Jennifer Rothberg2, Robert J. Gillies3 and Bonnie F. Sloane4 & Douglas Yingst 1Department of Physiology, 2Cancer Biology Program, and 4Department of Pharmacology, Wayne State University School of Medicine, Detroit, MI, 48201; 3H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33602 Breast cancer is the most commonly diagnosed cancer in women in USA and has a high mortality rate, second only to lung cancer. About 85% of total 63300 new cases of breast cancer are predicted to be ductal carcinoma in situ (DCIS) in 2012. We are interested in identifying markers that are predictive of changes that occur in the breast microenvironment as a result of the presence of premalignant lesions such as DCIS that are poised to develop into breast cancer. A critical barrier to cancer progression is its ability to survive in the acidic microenvironment characteristic of breast cancer. As the breast is comprised of different cell types, we performed gene expression analysis using Affymetrix gene chip HG U133 plus 2.0 array of 3 DCIS cell lines grown in 3D at neutral and acidic pH. We then computed the significantly changed genes at acidic pH for three DCIS cell lines and found 6 common and 121 similar genes. IPA core analysis of these genes revealed the interferon-signaling (IFN) pathway to be significantly altered. STAT1 was one key transcription factor that was upregulated and that might be driving downstream signaling as a response to acidic microenvironment. We are validating some of the downstream targets of the IFN pathway using qPCR. -

Alterations in the Intestinal Morphology, Gut Microbiota
nutrients Article Alterations in the Intestinal Morphology, Gut Microbiota, and Trace Mineral Status Following Intra-Amniotic Administration (Gallus gallus) of Teff (Eragrostis tef) Seed Extracts Johnathon Carboni 1, Spenser Reed 2,3, Nikolai Kolba 2 , Adi Eshel 4, Omry Koren 4 and Elad Tako 2,* 1 Department of Biological Sciences, Cornell University, Ithaca, NY 14853, USA; [email protected] 2 Department of Food Science, Cornell University, Stocking Hall, Ithaca, NY 14853-7201, USA; [email protected] (S.R.); [email protected] (N.K.) 3 Department of Family Medicine, Kaiser Permanente Fontana Medical Centers, Fontana, CA 92335, USA 4 Azrieli Faculty of Medicine, Bar-Ilan University, 1311502 Safed, Israel; [email protected] (A.E.); [email protected] (O.K.) * Correspondence: [email protected]; Tel.: +1-607-255-0884 Received: 20 August 2020; Accepted: 30 September 2020; Published: 2 October 2020 Abstract: The consumption of teff (Eragrostis tef ), a gluten-free cereal grain, has increased due to its dense nutrient composition including complex carbohydrates, unsaturated fatty acids, trace minerals (especially Fe), and phytochemicals. This study utilized the clinically-validated Gallus gallus intra amniotic feeding model to assess the effects of intra-amniotic administration of teff extracts versus controls using seven groups: (1) non-injected; (2) 18W H2O injected; (3) 5% inulin; (4) teff extract 1%; (5) teff extract 2.5%; (6) teff extract 5%; and (7) teff extract 7.5%. The treatment groups were compared to each other and to controls. Our data demonstrated a significant improvement in hepatic iron (Fe) and zinc (Zn) concentration and LA:DGLA ratio without concomitant serum concentration changes, up-regulation of various Fe and Zn brush border membrane proteins, and beneficial morphological changes to duodenal villi and goblet cells. -

Functional Characterisation of the Barley ZIP7 Zinc Transporter
Functional characterisation of the barley ZIP7 zinc transporter Jingwen Tiong B. Science (Hons), The University of Adelaide A thesis submitted for the degree of Doctor of Philosophy The University of Adelaide Faculty of Sciences School of Agriculture, Food & Wine Waite Campus February 2012 I Contents List of figures ......................................................................................................................... VI List of tables ......................................................................................................................... VII Abstract ............................................................................................................................... VIII Declaration .............................................................................................................................. X Acknowledgements ............................................................................................................... XI Glossary of abbreviations .................................................................................................. XIII Chapter 1: Literature Review ................................................................................................. 1 1.1 Introduction ...................................................................................................................... 1 1.2 Improvement of plant Zn nutrition and biofortification .................................................. 3 1.2.1 Application of Zn fertilisers as an agronomic -

Mirnas Documented to Play a Role in Hematopoietic Cell Lineage. Our
Table S1: miRNAs documented to play a role in hematopoietic cell lineage. Our review of the literature summarizing miRNAs known to be involved in the development and proliferation of the hematopoietic lineage cells. miRNA Expression/function/target/regulator References miR-150 Elevated during developmental stages of B and T cell maturation. 16-19 Controls B cell differentiation, present in mature, resting T and B cells but decreased upon activation of naïve T or B cells. Plays a role in establishing lymphocyte identity. Very little is known about function in T cells. Regulators: Foxp3 Target: C-Myb miR-146a/b Upregulated in macropgahe inflammatory response. Differentially 17, 20 upregulated in murine Th1 subset but abolished in Th2 subset. Upregulated in response to TCRs stimulation, as well as by IL-1 and TNF. Highly expressed in murine T-regs and could play a role in establishing lymphocyte identity. Modulates activation induced cell death in activated T cells. Negative regulator of TLR and cytokine signaling pathway. Endotoxin tolerance. Antiviral role. Targets: IRAK1, IRAK2, TRAF6, FAF1 miR-16-1 Promote apoptosis by targeting Bcl2 expression, act as tumor 22 cluster suppressor RNAs. May block differentiation of later stage hematopoietic progenitor cells to mature cells. Downregulated in CLL. Target: BCL2. miR-155 Regulator of T and B cell maturation and innate immune response. 23-29 Expressed in primary mediastinal B-cell lymphoma, T and B cells, macrophages and DCs. Upregulated during B cell activation. Involved in T cell differentiation and indicated as a positive regulator of cytokine production. Activated by stimulating TLR3 and INFab receptors in bone derived macrophages (regulation of antimicrobial defense). -

Genetic Variation of Traits Related to Salt Stress Response in Wheat (Triticum Aestivum L.)
Institut für Nutzpflanzenwissenschaften und Ressourcenschutz Genetic variation of traits related to salt stress response in Wheat (Triticum aestivum L.) Dissertation zur Erlangung des Grades Doktor der Agrarwissenschaften (Dr. agr.) der Landwirtschaftlichen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn vorgelegt von Oyiga Benedict Chijioke aus Enugu-Ezike, Nigeria Bonn 2017 Referent: Prof. Dr. Jens Léon Plant Breeding, INRES, University of Bonn Korreferent: Prof. Dr. Heiner Goldbach Plant Nutrition, INRES, University of Bonn Tag der Mündlichen Prüfung: 14.12. 2016 This research work was financially supported by the “Bundesministerium fur wirtschaftliche Zusammenarbeit und Entwicklung (BMZ)” in collaboration with the German Agency for International Cooperation (GIZ), Germany (Project number: 09.7860.1-001.00), the International Centre for Research in Dryland Agriculture (ICARDA) and the Center for Development Research (ZEF), Friedrich-Wilhelms-University, Bonn, Germany. Angefertigt mit Genehmigung der Landwirtschaftlichen Fakultät der Universität Bonn ii GENERAL SUMMARY (English) Salinity is one of the most severe abiotic stresses perceived by plants, and is continuously increasing due to climatic change and poor irrigation management practices. It is currently affecting ~800 million hectares of land worldwide, including over 20% of the world’s irrigated arable land. Salinity causes significant growth reduction and crop yield losses. With the predicted geometric increase in the global population, improving the salt tolerance (ST) of crops has become an important challenge and target for plant breeders. Several approaches have been exhaustively exploited to ameliorate the impact of salinity on crop plants, but because of the complex nature of ST in crop plant, these approaches have not been optimally translated into the desired results. -

Title Methods to Evaluate Zinc Transport Into and out of the Secretory and Endosomal-Lysosomal Compartments in DT40 Cells. Autho
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Kyoto University Research Information Repository Methods to Evaluate Zinc Transport into and out of the Title Secretory and Endosomal-Lysosomal Compartments in DT40 Cells. Author(s) Kambe, Taiho Citation Methods in enzymology (2014), 534: 77-92 Issue Date 2014 URL http://hdl.handle.net/2433/180109 Right © 2014 Elsevier Inc. Type Journal Article Textversion author Kyoto University Running title: Zinc transport in subcellular compartments Methods to evaluate zinc transport into and out of the secretory and endosomal-lysosomal compartments in DT40 cells Taiho Kambe Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan Email: [email protected] Tel: +81-75-753-6273 Keywords: zinc, ZIP (Zrt/Irt-like protein), ZnT (zinc transporter), signaling, secretory and endosomal-lysosomal compartments, DT40, multiple gene disruptions 1 Abstract Zinc plays crucial roles in diverse biological processes. Recently, in addition to zinc mobilization into and out of the cell, zinc mobilization into and out of intracellular organelles, including the secretory and endosomal-lysosomal compartments, has received growing interest. In vertebrate cells, the Zrt/Irt-like proteins (ZIPs) and zinc transporters (ZnTs) are the two major families of zinc transporters involved in zinc mobilization across cellular membranes. Importantly, nearly half of ZIP and ZnT transporters are localized to subcellular compartments. Thus, to elucidate the numerous zinc-related cellular events, understanding ZIP and ZnT functions is critical. This chapter describes advanced methods used in our laboratory to examine zinc mobilization. Specifically, genetic and molecular approaches using chicken DT40 cells deficient in multiple ZnT and ZIP transporters are described. -

The Function of Zinc Transporters in Human Cells
THE FUNCTION OF ZINC TRANSPORTERS IN HUMAN CELLS BY LOVELEEN KUMAR, BSc., MSc. Submitted in fulfilment of requirements for the degree of Doctor of Philosophy DEAKIN UNIVERSITY JULY 2014 i Acknowledgements I would sincerely like to thank my supervisor Professor Leigh Ackland for giving me the opportunity to undertake my PhD under her invaluable guidance with continual support, encouragement and infinite enthusiasm. Thank you so much Leigh for having faith in me and being wonderful supervisor one could ever wish for. I would like to thank my co-supervisor Dr Agnes Michalczyk, not only for providing invaluable supervision and encouragement but being a great friend who I could talk to during hard times. Thanks a lot Agnes for being always there to support me. I am grateful to Professor Julian Mercer for allowing me to undertake my PhD candidature in the CCMB. I would also like to thank him for his expert advice at the discussion sessions that made me think in detail. Special thanks to Prof Andy Sinclair for providing me the scholarship and the mentoring sessions we had, it made me concentrate on the main goal. I would like to thank my Lab Manager Mr. Michael Holmes for having faith in my capability and providing me with opportunity to work as a technical officer to financially support my family. I am very grateful to our school manager Marita Reynolds to sanction my study leave to finish off my writting. I would like to extend that thanks to all Technical staff, who gave enormous mental support and friendly environment that helped me come out of my grief. -

Supporting Information
Supporting Information Redefining the breast cancer exosome proteome by tandem mass tag quantitative proteomics and multivariate cluster analysis. David J Clarkǂ*1,2, William E Fondrieǂ2,3, Zhongping Liao4, Phyllis I Hanson5, Amy Fulton2, Li Mao1,2, Austin J Yang2 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, Baltimore, MD 2 Marlene and Stewart Greenebaum Cancer Center, University of Maryland, Baltimore, MD 3Center for Vascular and Inflammatory Diseases, University of Maryland School of Medicine, Baltimore, MD 4 Lily Research Laboratory, Eli Lily and Company, Indianapolis, IN 5Department of Cell Biology and Physiology, Washington University, St. Louis, MO ǂ These authors contributed equally to this work. *Corresponding author: [email protected] 410-706-6114 Table of Contents Figure S-1: TMT log2 protein ratios were plotted by 100K enrichment against Optiprep/100K enrichment. (pg. S-2) Figure S-2: Immunoblot of SEC fractions, and Venn diagram of SEC and TMT-SVM analysis. (pg. S-3) Table S-1 Protein identifications, TMT ratios, SVM Classification and SVM Probability of 10K, 100K, and Optiprep lysates derived from SKBR3B conditioned media. (pg. S-4) Table S-2. Plasma membrane markers and their respective SVM Classification (pg. S-75) Table S-3. Identified proteins from our SEC analysis with respective Protein Prophet probability (pg. S-76) S-1 Figure S-1. Known exosome markers localize to quadrant corresponding to 100K enrichment. TMT log2 protein ratios were plotted by 100K enrichment (x-axis) against Optiprep/100K enrichment (y-axis). Selected Exosome (red) and Non-exosome (blue) protein markers are annotated. -

Zinc Signaling in Physiology and Pathogenesis
International Journal of Molecular Sciences Books Zinc Signaling in Physiology and Pathogenesis Edited by Toshiyuki Fukada and Taiho Kambe Printed Edition of the Special Issue Published in IJMS www.mdpi.com/journal/ijms MDPI Zinc Signaling in Physiology and Pathogenesis Special Issue Editors Toshiyuki Fukada Books Taiho Kambe MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade MDPI Special Issue Editors ToshiyukiFukada TokushimaBunri University Japan TaihoKambe Kyoto University Japan Editorial Office MDPI St. Alban-Anlage 66 Basel, Switzerland This edition is a reprint of the Special Issue published online in the open access journal IJMS (ISSN 1422-0067) from 2017-2018(available at: http:/ /www.mdpi.com/journal/ijms/special...issues/ zinc...signaling). Books For citation purposes, cite each article independently as indicated on the article page online and as indicated below: Lastname, F.M.; Lastname, F.M. Article title. Journal Name Year,A rticle number, page range. First Edition 2018 ISBN 978-3-03842-821-3 (Pbk) ISBN 978-3-03842-822-0 (PDF) Cover photo courtesy ofKoh-ei Toyoshimaand TakashiTsuji. It depicts mouse epidermal hair follicles which express zinc transporter ZIPlO (Bum-Ho Bin et al. PNAS 114, 12243-12248, 2017) Articles in this volume are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles even for commercial purposes, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book taken as a whole is©2018 MDPI, Basel, Switzerland, distributed under the terms and conditions of the Creative Commons license CC BY-NC-ND(http://creativecommons.org/licenses/by-nc-nd/ 4.0/).