Fossils, Phylogeny, and Anatomical Regions: Insights Exemplified Through Turtles
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Xenosaurus Tzacualtipantecus. the Zacualtipán Knob-Scaled Lizard Is Endemic to the Sierra Madre Oriental of Eastern Mexico
Xenosaurus tzacualtipantecus. The Zacualtipán knob-scaled lizard is endemic to the Sierra Madre Oriental of eastern Mexico. This medium-large lizard (female holotype measures 188 mm in total length) is known only from the vicinity of the type locality in eastern Hidalgo, at an elevation of 1,900 m in pine-oak forest, and a nearby locality at 2,000 m in northern Veracruz (Woolrich- Piña and Smith 2012). Xenosaurus tzacualtipantecus is thought to belong to the northern clade of the genus, which also contains X. newmanorum and X. platyceps (Bhullar 2011). As with its congeners, X. tzacualtipantecus is an inhabitant of crevices in limestone rocks. This species consumes beetles and lepidopteran larvae and gives birth to living young. The habitat of this lizard in the vicinity of the type locality is being deforested, and people in nearby towns have created an open garbage dump in this area. We determined its EVS as 17, in the middle of the high vulnerability category (see text for explanation), and its status by the IUCN and SEMAR- NAT presently are undetermined. This newly described endemic species is one of nine known species in the monogeneric family Xenosauridae, which is endemic to northern Mesoamerica (Mexico from Tamaulipas to Chiapas and into the montane portions of Alta Verapaz, Guatemala). All but one of these nine species is endemic to Mexico. Photo by Christian Berriozabal-Islas. amphibian-reptile-conservation.org 01 June 2013 | Volume 7 | Number 1 | e61 Copyright: © 2013 Wilson et al. This is an open-access article distributed under the terms of the Creative Com- mons Attribution–NonCommercial–NoDerivs 3.0 Unported License, which permits unrestricted use for non-com- Amphibian & Reptile Conservation 7(1): 1–47. -
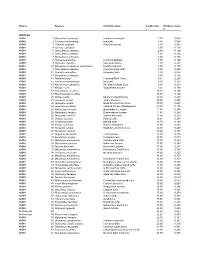
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

The PARI Journal Vol. VI, No. 3
ThePARIJournal A quarterly publication of the Pre-Columbian Art Research Institute Volume VI, No.3, Winter 2006 Teasing the Turtle from its Shell: In This Issue: AHKand MAHK in MayaWriting* Teasing the Turtle MARC ZENDER from its Shell: Peabody Museum of Archaeology & Ethnology, Harvard University AHK and MAHK in Maya Writing resents the profile carapace of a testudine by Marc Zender (i.e., a turtle, tortoise or terrapin), careful PAGES 1-14 examination of their formal features, dis- tributions and lexical associations reveals • them to have been quite distinct entities, Morley's Diary: rigidly distinguished by scribes through- a b out the life of the script. As discussed in April 23, 1932 detail below, there is in fact strong reason Figure 1. “Turtle Shell” Glyphs: (a) MAHK “carapace” and PAGES 15-16 “cover”; (b) AHK “turtle.” All drawings by the author. to see the first sign (Figure 1a) as the de- piction of a turtle shell carrying the value Formally similar but nevertheless dis- MAHK “carapace, shell,” and, less specifi- tinct signs remain a constant challenge to cally, “cover, enclosure.” By contrast, the Joel Skidmore students of the ancient Maya script. Pre- latter sign (Figure 1b), while also depict- Editor ing a turtle shell, has long been known [email protected] viously I have argued that a number of signs traditionally considered identical to carry the value AHK “turtle,” and is are in fact quite distinct, as demonstrated therefore best thought of as the pars pro The PARI Journal by formal divergences and consistent dis- toto depiction of a “turtle” proper. -

Terrapene Carolina (Linnaeus 1758) – Eastern Box Turtle, Common Box Turtle
Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project ofEmydidae the IUCN/SSC — TortoiseTerrapene and Freshwatercarolina Turtle Specialist Group 085.1 A.G.J. Rhodin, P.C.H. Pritchard, P.P. van Dijk, R.A. Saumure, K.A. Buhlmann, J.B. Iverson, and R.A. Mittermeier, Eds. Chelonian Research Monographs (ISSN 1088-7105) No. 5, doi:10.3854/crm.5.085.carolina.v1.2015 © 2015 by Chelonian Research Foundation • Published 26 January 2015 Terrapene carolina (Linnaeus 1758) – Eastern Box Turtle, Common Box Turtle A. ROSS KIESTER1 AND LISABETH L. WILLEY2 1Turtle Conservancy, 49 Bleecker St., Suite 601, New York, New York 10012 USA [[email protected]]; 2Department of Environmental Studies, Antioch University New England, 40 Avon St., Keene, New Hampshire 03431 USA [[email protected]] SUMMARY. – The Eastern Box Turtle, Terrapene carolina (Family Emydidae), as currently understood, contains six living subspecies of small turtles (carapace lengths to ca. 115–235 mm) able to close their hinged plastrons into a tightly closed box. Although the nominate subspecies is among the most widely distributed and well-known of the world’s turtles, the two Mexican subspecies are poorly known. This primarily terrestrial, though occasionally semi-terrestrial, species ranges throughout the eastern and southern United States and disjunctly in Mexico. It was generally recognized as common in the USA throughout the 20th century, but is now threatened by continuing habitat conversion, road mortality, and collection for the pet trade, and notable population declines have been documented throughout its range. In the United States, this turtle is a paradigm example of the conservation threats that beset and impact a historically common North American species. -

A Systematic Review of the Turtle Family Emydidae
67 (1): 1 – 122 © Senckenberg Gesellschaft für Naturforschung, 2017. 30.6.2017 A Systematic Review of the Turtle Family Emydidae Michael E. Seidel1 & Carl H. Ernst 2 1 4430 Richmond Park Drive East, Jacksonville, FL, 32224, USA and Department of Biological Sciences, Marshall University, Huntington, WV, USA; [email protected] — 2 Division of Amphibians and Reptiles, mrc 162, Smithsonian Institution, P.O. Box 37012, Washington, D.C. 200137012, USA; [email protected] Accepted 19.ix.2016. Published online at www.senckenberg.de / vertebrate-zoology on 27.vi.2016. Abstract Family Emydidae is a large and diverse group of turtles comprised of 50 – 60 extant species. After a long history of taxonomic revision, the family is presently recognized as a monophyletic group defined by unique skeletal and molecular character states. Emydids are believed to have originated in the Eocene, 42 – 56 million years ago. They are mostly native to North America, but one genus, Trachemys, occurs in South America and a second, Emys, ranges over parts of Europe, western Asia, and northern Africa. Some of the species are threatened and their future survival depends in part on understanding their systematic relationships and habitat requirements. The present treatise provides a synthesis and update of studies which define diversity and classification of the Emydidae. A review of family nomenclature indicates that RAFINESQUE, 1815 should be credited for the family name Emydidae. Early taxonomic studies of these turtles were based primarily on morphological data, including some fossil material. More recent work has relied heavily on phylogenetic analyses using molecular data, mostly DNA. The bulk of current evidence supports two major lineages: the subfamily Emydinae which has mostly semi-terrestrial forms ( genera Actinemys, Clemmys, Emydoidea, Emys, Glyptemys, Terrapene) and the more aquatic subfamily Deirochelyinae ( genera Chrysemys, Deirochelys, Graptemys, Malaclemys, Pseudemys, Trachemys). -

Chelonian Advisory Group Regional Collection Plan 4Th Edition December 2015
Association of Zoos and Aquariums (AZA) Chelonian Advisory Group Regional Collection Plan 4th Edition December 2015 Editor Chelonian TAG Steering Committee 1 TABLE OF CONTENTS Introduction Mission ...................................................................................................................................... 3 Steering Committee Structure ........................................................................................................... 3 Officers, Steering Committee Members, and Advisors ..................................................................... 4 Taxonomic Scope ............................................................................................................................. 6 Space Analysis Space .......................................................................................................................................... 6 Survey ........................................................................................................................................ 6 Current and Potential Holding Table Results ............................................................................. 8 Species Selection Process Process ..................................................................................................................................... 11 Decision Tree ........................................................................................................................... 13 Decision Tree Results ............................................................................................................. -

Turtles of the World, 2010 Update: Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status
Conservation Biology of Freshwater Turtles and Tortoises: A Compilation ProjectTurtles of the IUCN/SSC of the World Tortoise – 2010and Freshwater Checklist Turtle Specialist Group 000.85 A.G.J. Rhodin, P.C.H. Pritchard, P.P. van Dijk, R.A. Saumure, K.A. Buhlmann, J.B. Iverson, and R.A. Mittermeier, Eds. Chelonian Research Monographs (ISSN 1088-7105) No. 5, doi:10.3854/crm.5.000.checklist.v3.2010 © 2010 by Chelonian Research Foundation • Published 14 December 2010 Turtles of the World, 2010 Update: Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status TUR T LE TAXONOMY WORKING GROUP * *Authorship of this article is by this working group of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, which for the purposes of this document consisted of the following contributors: ANDERS G.J. RHODIN 1, PE T ER PAUL VAN DI J K 2, JOHN B. IVERSON 3, AND H. BRADLEY SHAFFER 4 1Chair, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Chelonian Research Foundation, 168 Goodrich St., Lunenburg, Massachusetts 01462 USA [[email protected]]; 2Deputy Chair, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, Virginia 22202 USA [[email protected]]; 3Department of Biology, Earlham College, Richmond, Indiana 47374 USA [[email protected]]; 4Department of Evolution and Ecology, University of California, Davis, California 95616 USA [[email protected]] AB S T RAC T . – This is our fourth annual compilation of an annotated checklist of all recognized and named taxa of the world’s modern chelonian fauna, documenting recent changes and controversies in nomenclature, and including all primary synonyms, updated from our previous three checklists (Turtle Taxonomy Working Group [2007b, 2009], Rhodin et al. -

BMC Evolutionary Biology Biomed Central
BMC Evolutionary Biology BioMed Central Research article Open Access Assessing what is needed to resolve a molecular phylogeny: simulations and empirical data from emydid turtles Phillip Q Spinks*1,2, Robert C Thomson1,2, Geoff A Lovely1,3 and H Bradley Shaffer1,2 Address: 1Department of Evolution and Ecology, Davis, USA, 2Center for Population Biology, University of California, Davis, USA and 3Present address Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, USA Email: Phillip Q Spinks* - [email protected]; Robert C Thomson - [email protected]; Geoff A Lovely - [email protected]; H Bradley Shaffer - [email protected] * Corresponding author Published: 12 March 2009 Received: 24 July 2008 Accepted: 12 March 2009 BMC Evolutionary Biology 2009, 9(56): 1-17 © 2009 Spinks et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Phylogenies often contain both well-supported and poorly supported nodes. Determining how much additional data might be required to eventually recover most or all nodes with high support is an important pragmatic goal, and simulations have been used to examine this question. Most simulations have been based on few empirical loci, and suggest that well supported phylogenies can be determined with a very modest amount of data. Here we report the results of an empirical phylogenetic analysis of all 10 genera and 25 of 48 species of the new world pond turtles (family Emydidae) based on one mitochondrial (1070 base pairs) and seven nuclear loci (5961 base pairs), and a more biologically realistic simulation analysis incorporating variation among gene trees, aimed at determining how much more data might be necessary to recover weakly-supported nodes with strong support. -

Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status
Conservation Biology of Freshwater Turtles and Tortoises: A Compilation ProjectTurtles of the IUCN/SSC of the World Tortoise – 2012and Freshwater Checklist Turtle Specialist Group 000.243 A.G.J. Rhodin, P.C.H. Pritchard, P.P. van Dijk, R.A. Saumure, K.A. Buhlmann, J.B. Iverson, and R.A. Mittermeier, Eds. Chelonian Research Monographs (ISSN 1088-7105) No. 5, doi:10.3854/crm.5.000.checklist.v5.2012 © 2012 by Chelonian Research Foundation • Published 31 December 2012 Turtles of the World, 2012 Update: Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status TUR T LE TAXONOMY WORKING GROUP * *Authorship of this article is by this working group of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, which for the purposes of this document consisted of the following contributors: PE T ER PAUL VAN DIJK 1, JOHN B. IVERSON 2, H. BRA D LEY SHAFFER 3, ROGER BOUR 4, AN D AN D ERS G.J. RHO D IN 5 1Co-Chair, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, Virginia 22202 USA [[email protected]]; 2Department of Biology, Earlham College, Richmond, Indiana 47374 USA [[email protected]]; 3Department of Ecology and Evolutionary Biology and Institute of the Environment and Sustainability, University of California, Los Angeles, California 90095 USA [[email protected]]; 4Laboratoire des Reptiles et Amphibiens, Muséum National d’Histoire Naturelle, Paris, France [[email protected]]; 5Chairman Emeritus, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Chelonian Research Foundation, 168 Goodrich St., Lunenburg, Massachusetts 01462 USA [[email protected]] AB S T RAC T . -

Box Turtle Connection–Building a Legacy
THEa BOX TURTLE CONNECTION a THE BOX TURTLE CONNECTION Building a Legacy Building a Legacy Ann Berry Somers, Catherine E. Matthews, and Ashley A. LaVere Ann Berry Somers, Catherine E. Matthews, and Ashley A. LaVere Published 2017 Cover image by J.D. Willson, male Terrapene carolina carolina from Aiken County, South Carolina. 2006. Our work on this second edition of the book has been partially funded by an informal science education National Science Foundation grant (DRL-1114558), Herpetology Education in Rural Places & Spaces (The HERP Project) Any opinions, findings, conclusions or recommendations expressed do not necessarily reflect the views of the National Science Foundation. The Box Turtle Connection Building a Legacy We dedicate this book to The Box Turtle Connection Project Leaders. They are the foundation on which this project is built. Their hard work and passion act as the frame that shapes and supports this long-term study. With their help, we will continue to further our knowledge about box turtles and strive to protect them and their habitats. Mike Vaughan https://boxturtle.uncg.edu/ “To the amateur, I say learn about nature, but do it carefully and with commitment . Amateur naturalists have much to offer the scientific community in understanding the ways of box turtles. To the professional, I say get outdoors and observe nature . Natural history should be celebrated. After all, we are dealing with life in all its complexity. A study of box turtles is the perfect place to begin.” Ken Dodd (2001) Contents Contents .......................................................................................................................................... -
A Prehispanic Maya Katun Wheel
CHAPTER 2 A Prehispanic Maya Katun Wheel From the beginnings of the Classic period to the mid-eighteenth century, a span of some twelve hundred years, the Katun has been one of the basic time units of the Maya. Composed of twenty Tuns of 360 days, the Katun is almost twenty years, to be precise, 7,200 days. In Classic period inscriptions it is but the fourth of five time units composing a continual count of days from a mythical event in 3114 BC. But the Katun is a great deal more than a dull cog of Maya calendrics; it constitutes an essential element of traditional Maya history and reli- gion. The great majority of Classic Maya monuments commemorate the ending of the Katun period. Classic Maya lords proclaimed the number of Katun endings experienced during their lifetimes as a sort of title. In Postclassic and colonial Yucatan, invasions, droughts, and even the creation and destruction of the world were recorded and foretold in terms of the Katun cycle. But although the progress and completion of the Katun is expressed repeatedly in prehispanic and colonial Maya accounts, we have little understanding about how the passage of the Katun periods was actually perceived. The focus of this study is upon the succession of Katuns of Postclassic and colonial Yucatan. I will demonstrate that the turtle was an important means of describing the Katun cycle. In both the Classic and Postclassic periods, this creature was explicitly identified with period ending dates. Moreover, the Postclassic data provide strong evidence for the importance of penitential bloodletting at period ending ceremonies. -

Support the Legal and Sustainable Trade in Turtles and Tortoises
Trinational Trade and Enforcement Training Workshop to Support the Legal and Sustainable Trade in Turtles and Tortoises Commission for Environmental Cooperation – August 2019 Trinational Trade and Enforcement Training Workshop to Support the Legal and Sustainable Trade in Turtles and Tortoises i Please cite as: CEC. 2019. Trinational Trade and Enforcement Training Workshop to Support the Legal and Sustainable Trade in Turtles and Tortoises. Report. Montreal, Canada: Commission for Environmental Cooperation. 2019. 84 pp. This publication was prepared by Kurt A. Buhlmann, Michael J. Dreslik, and Peter Paul van Dijk for the Secretariat of the Commission for Environmental Cooperation. The information contained herein is the responsibility of the author and does not necessarily reflect the views of the CEC or the governments of Canada, Mexico or the United States of America. About the author(s): Kurt A. Buhlmann, Ph.D., is a conservation ecologist, focusing on recovery of threatened amphibians and reptiles, and operates Buhlmann Ecological Research and Consulting, LLC, and is also a Senior Research Associate at the University of Georgia’s Savannah River Ecology Laboratory. Michael J. Dreslik, Ph.D., is an ecologist with Applied Envirolytics, LLC. Peter Paul van Dijk, Ph.D., is Director of Turtle Conservation Programs, with Global Wildlife Conservation; Field Conservation Director at the Turtle Conservancy; and deputy chair of the IUCN SSC Tortoise & Freshwater Turtle Specialist Group. Reproduction of this document in whole or in part and in any form for educational or non-profit purposes may be made without special permission from the CEC Secretariat, provided acknowledgment of the source is made. The CEC would appreciate receiving a copy of any publication or material that uses this document as a source.