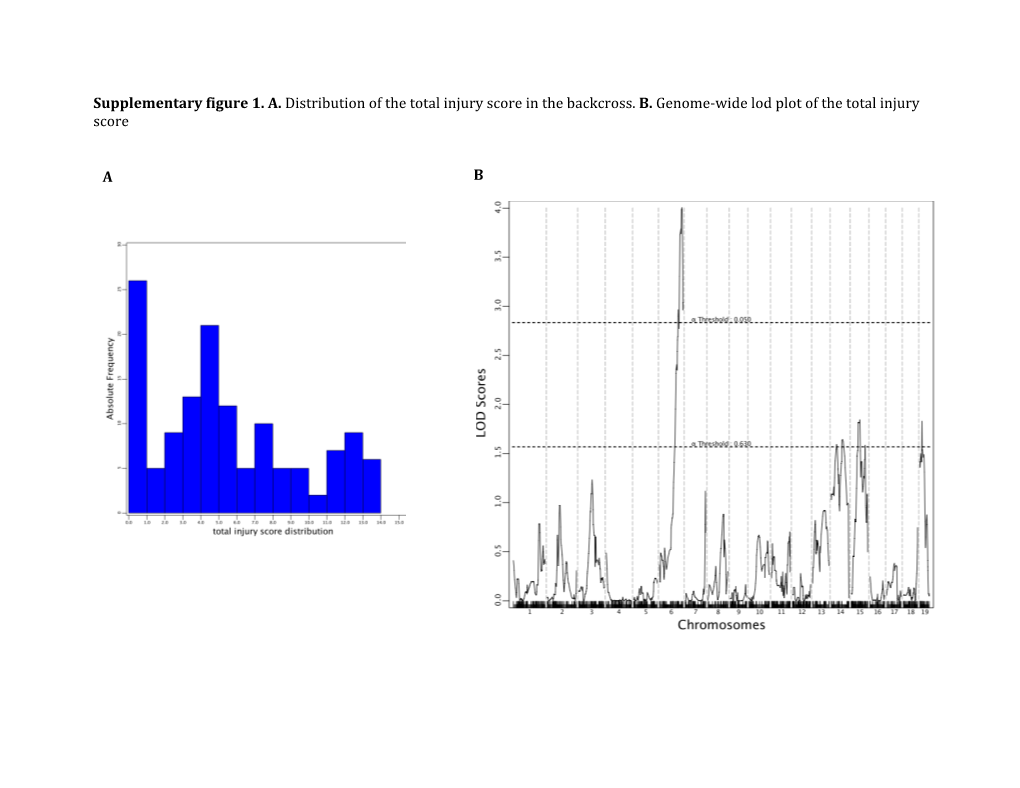
Supplementary Figure 1. A. Distribution of the Total Injury Score in the Backcross
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Down-Regulation of Stem Cell Genes, Including Those in a 200-Kb Gene Cluster at 12P13.31, Is Associated with in Vivo Differentiation of Human Male Germ Cell Tumors
Research Article Down-Regulation of Stem Cell Genes, Including Those in a 200-kb Gene Cluster at 12p13.31, Is Associated with In vivo Differentiation of Human Male Germ Cell Tumors James E. Korkola,1 Jane Houldsworth,1,2 Rajendrakumar S.V. Chadalavada,1 Adam B. Olshen,3 Debbie Dobrzynski,2 Victor E. Reuter,4 George J. Bosl,2 and R.S.K. Chaganti1,2 1Cell Biology Program and Departments of 2Medicine, 3Epidemiology and Biostatistics, and 4Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York Abstract on the degree and type of differentiation (i.e., seminomas, which Adult male germ cell tumors (GCTs) comprise distinct groups: resemble undifferentiated primitive germ cells, and nonseminomas, seminomas and nonseminomas, which include pluripotent which show varying degrees of embryonic and extraembryonic embryonal carcinomas as well as other histologic subtypes patterns of differentiation; refs. 2, 3). Nonseminomatous GCTs are exhibiting various stages of differentiation. Almost all GCTs further subdivided into embryonal carcinomas, which show early show 12p gain, but the target genes have not been clearly zygotic or embryonal-like differentiation, yolk sac tumors and defined. To identify 12p target genes, we examined Affymetrix choriocarcinomas, which exhibit extraembryonal forms of differ- (Santa Clara, CA) U133A+B microarray (f83% coverage of 12p entiation, and teratomas, which show somatic differentiation along genes) expression profiles of 17 seminomas, 84 nonseminoma multiple lineages (3). Both seminomas and embryonal carcinoma GCTs, and 5 normal testis samples. Seventy-three genes on 12p are known to express stem cell markers, such as POU5F1 (4) and were significantly overexpressed, including GLUT3 and REA NANOG (5). -

Datasheet PB1029 Anti-AEBP2 Antibody
Product datasheet Anti-AEBP2 Antibody Catalog Number: PB1029 BOSTER BIOLOGICAL TECHNOLOGY Special NO.1, International Enterprise Center, 2nd Guanshan Road, Wuhan, China Web: www.boster.com.cn Phone: +86 27 67845390 Fax: +86 27 67845390 Email: [email protected] Basic Information Product Name Anti-AEBP2 Antibody Gene Name AEBP2 Source Rabbit IgG Species Reactivity human,mouse,rat Tested Application WB,IHC-P,ICC/IF,FCM Contents 500ug/ml antibody with PBS ,0.02% NaN3 , 1mg BSA and 50% glycerol. Immunogen E.coli-derived human AEBP2 recombinant protein (Position: K424-Q517). Human AEBP2 shares 98.8% amino acid (aa) sequence identity with mouse AEBP2. Purification Immunogen affinity purified. Observed MW 54KD Dilution Ratios Western blot: 1:500-2000 Immunohistochemistry(Paraffin-embedded Section): 1:50-400 Immunocytochemistry/Immunofluorescence (ICC/IF): 1:50-400 Flow cytometry (FCM): 1-3μg/1x106 cells Storage 12 months from date of receipt,-20℃ as supplied.6 months 2 to 8℃ after reconstitution. Avoid repeated freezing and thawing Background Information Adipocyte Enhancer-Binding Protein is a zinc finger protein that in humans is encoded by the evolutionarily well-conserved gene AEBP2. This gene is mapped to 12p12.3. AEBP2 is a DNA-binding transcriptional repressor. It may regulate the migration and development of the neural crest cells through the PRC2-mediated epigenetic mechanism and is most likely a targeting protein for the mammalian PRC2 complex. Reference Anti-AEBP2 Antibody被引用在0文献中。 暂无引用 FOR RESEARCH USE ONLY. NOT FOR DIAGNOSTIC AND CLINICAL USE. 1 Product datasheet Anti-AEBP2 Antibody Catalog Number: PB1029 BOSTER BIOLOGICAL TECHNOLOGY Special NO.1, International Enterprise Center, 2nd Guanshan Road, Wuhan, China Web: www.boster.com.cn Phone: +86 27 67845390 Fax: +86 27 67845390 Email: [email protected] Selected Validation Data Figure 1. -

Identification of Chebulinic Acid As a Dual Targeting Inhibitor of Protein
Bioorganic Chemistry 90 (2019) 103087 Contents lists available at ScienceDirect Bioorganic Chemistry journal homepage: www.elsevier.com/locate/bioorg Short communication Identification of chebulinic acid as a dual targeting inhibitor of protein T tyrosine phosphatases relevant to insulin resistance Sun-Young Yoona,1, Hyo Jin Kangb,1, Dohee Ahna, Ji Young Hwanga, Se Jeong Kwona, ⁎ Sang J. Chunga, a School of Pharmacy, Sungkyunkwan University, Suwon 16419, Republic of Korea b Department of Chemistry, Dongguk University, Seoul 100-715, Republic of Korea ARTICLE INFO ABSTRACT Keywords: Natural products as antidiabetic agents have been shown to stimulate insulin signaling via the inhibition of the Protein tyrosine phosphatases (PTPs) protein tyrosine phosphatases relevant to insulin resistance. Previously, we have identified PTPN9 and DUSP9 as Chebulinic acid potential antidiabetic targets and a multi-targeting natural product thereof. In this study, knockdown of PTPN11 Type 2 diabetes increased AMPK phosphorylation in differentiated C2C12 muscle cells by 3.8 fold, indicating that PTPN11 could Glucose-uptake be an antidiabetic target. Screening of a library of 658 natural products against PTPN9, DUSP9, or PTPN11 PTPN9 identified chebulinic acid (CA) as a strong allosteric inhibitor with a slow cooperative binding toPTPN9 PTPN11 (IC50 = 34 nM) and PTPN11 (IC50 = 37 nM), suggesting that it would be a potential antidiabetic candidate. Furthermore, CA stimulated glucose uptake and resulted in increased AMP-activated protein kinase (AMPK) phosphorylation. Taken together, we demonstrated that CA increased glucose uptake as a dual inhibitor of PTPN9 and PTPN11 through activation of the AMPK signaling pathway. These results strongly suggest that CA could be used as a potential therapeutic candidate for the treatment of type 2 diabetes. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Glucocorticoid Receptor Signaling Activates TEAD4 to Promote Breast
Published OnlineFirst July 9, 2019; DOI: 10.1158/0008-5472.CAN-19-0012 Cancer Molecular Cell Biology Research Glucocorticoid Receptor Signaling Activates TEAD4 to Promote Breast Cancer Progression Lingli He1,2, Liang Yuan3,Yang Sun1,2, Pingyang Wang1,2, Hailin Zhang4, Xue Feng1,2, Zuoyun Wang1,2, Wenxiang Zhang1,2, Chuanyu Yang4,Yi Arial Zeng1,2,Yun Zhao1,2,3, Ceshi Chen4,5,6, and Lei Zhang1,2,3 Abstract The Hippo pathway plays a critical role in cell growth and to the TEAD4 promoter to boost its own expression. Func- tumorigenesis. The activity of TEA domain transcription factor tionally, the activation of TEAD4 by GC promoted breast 4 (TEAD4) determines the output of Hippo signaling; how- cancer stem cells maintenance, cell survival, metastasis, and ever, the regulation and function of TEAD4 has not been chemoresistance both in vitro and in vivo. Pharmacologic explored extensively. Here, we identified glucocorticoids (GC) inhibition of TEAD4 inhibited GC-induced breast cancer as novel activators of TEAD4. GC treatment facilitated gluco- chemoresistance. In conclusion, our study reveals a novel corticoid receptor (GR)-dependent nuclear accumulation and regulation and functional role of TEAD4 in breast cancer and transcriptional activation of TEAD4. TEAD4 positively corre- proposes a potential new strategy for breast cancer therapy. lated with GR expression in human breast cancer, and high expression of TEAD4 predicted poor survival of patients with Significance: This study provides new insight into the role breast cancer. Mechanistically, GC activation promoted GR of glucocorticoid signaling in breast cancer, with potential for interaction with TEAD4, forming a complex that was recruited clinical translation. -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -

Full Text (PDF)
Published OnlineFirst May 1, 2014; DOI: 10.1158/0008-5472.CAN-13-3147 Cancer Tumor and Stem Cell Biology Research Loss of the Polycomb Mark from Bivalent Promoters Leads to Activation of Cancer-Promoting Genes in Colorectal Tumors Maria A. Hahn1, Arthur X. Li2, Xiwei Wu3, Richard Yang1, David A. Drew4, Daniel W. Rosenberg4, and Gerd P. Pfeifer1 Abstract In colon tumors, the transcription of many genes becomes deregulated by poorly defined epigenetic mechanisms that have been studied mainly in established cell lines. In this study, we used frozen human colon tissues to analyze patterns of histone modification and DNA cytosine methylation in cancer and matched normal mucosa specimens. DNA methylation is strongly targeted to bivalent H3K4me3- and H3K27me3-associated promoters, which lose both histone marks and acquire DNA methylation. However, we found that loss of the Polycomb mark H3K27me3 from bivalent promoters was accompanied often by activation of genes associated with cancer progression, including numerous stem cell regulators, oncogenes, and proliferation-associated genes. Indeed, we found many of these same genes were also activated in patients with ulcerative colitis where chronic inflammation predisposes them to colon cancer. Based on our findings, we propose that a loss of Polycomb repression at bivalent genes combined with an ensuing selection for tumor-driving events plays a major role in cancer progression. Cancer Res; 74(13); 3617–29. Ó2014 AACR. Introduction cells (21, 22). The acquisition of the more permanent silencing Tumorigenesis is a complex process that is driven by a mark, 5mC, at the promoters of Polycomb target genes does number of genetic and epigenetic alterations, which often not fundamentally change their expression levels although result in aberrant gene expression (1–3). -

Accompanies CD8 T Cell Effector Function Global DNA Methylation
Global DNA Methylation Remodeling Accompanies CD8 T Cell Effector Function Christopher D. Scharer, Benjamin G. Barwick, Benjamin A. Youngblood, Rafi Ahmed and Jeremy M. Boss This information is current as of October 1, 2021. J Immunol 2013; 191:3419-3429; Prepublished online 16 August 2013; doi: 10.4049/jimmunol.1301395 http://www.jimmunol.org/content/191/6/3419 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130139 Material 5.DC1 References This article cites 81 articles, 25 of which you can access for free at: http://www.jimmunol.org/content/191/6/3419.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 1, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Global DNA Methylation Remodeling Accompanies CD8 T Cell Effector Function Christopher D. Scharer,* Benjamin G. Barwick,* Benjamin A. Youngblood,*,† Rafi Ahmed,*,† and Jeremy M. -

The Regulatory Roles of Phosphatases in Cancer
Oncogene (2014) 33, 939–953 & 2014 Macmillan Publishers Limited All rights reserved 0950-9232/14 www.nature.com/onc REVIEW The regulatory roles of phosphatases in cancer J Stebbing1, LC Lit1, H Zhang, RS Darrington, O Melaiu, B Rudraraju and G Giamas The relevance of potentially reversible post-translational modifications required for controlling cellular processes in cancer is one of the most thriving arenas of cellular and molecular biology. Any alteration in the balanced equilibrium between kinases and phosphatases may result in development and progression of various diseases, including different types of cancer, though phosphatases are relatively under-studied. Loss of phosphatases such as PTEN (phosphatase and tensin homologue deleted on chromosome 10), a known tumour suppressor, across tumour types lends credence to the development of phosphatidylinositol 3--kinase inhibitors alongside the use of phosphatase expression as a biomarker, though phase 3 trial data are lacking. In this review, we give an updated report on phosphatase dysregulation linked to organ-specific malignancies. Oncogene (2014) 33, 939–953; doi:10.1038/onc.2013.80; published online 18 March 2013 Keywords: cancer; phosphatases; solid tumours GASTROINTESTINAL MALIGNANCIES abs in sera were significantly associated with poor survival in Oesophageal cancer advanced ESCC, suggesting that they may have a clinical utility in Loss of PTEN (phosphatase and tensin homologue deleted on ESCC screening and diagnosis.5 chromosome 10) expression in oesophageal cancer is frequent, Cao et al.6 investigated the role of protein tyrosine phosphatase, among other gene alterations characterizing this disease. Zhou non-receptor type 12 (PTPN12) in ESCC and showed that PTPN12 et al.1 found that overexpression of PTEN suppresses growth and protein expression is higher in normal para-cancerous tissues than induces apoptosis in oesophageal cancer cell lines, through in 20 ESCC tissues. -

Polycomb Repressor Complex 2 Function in Breast Cancer (Review)
INTERNATIONAL JOURNAL OF ONCOLOGY 57: 1085-1094, 2020 Polycomb repressor complex 2 function in breast cancer (Review) COURTNEY J. MARTIN and ROGER A. MOOREHEAD Department of Biomedical Sciences, Ontario Veterinary College, University of Guelph, Guelph, ON N1G2W1, Canada Received July 10, 2020; Accepted September 7, 2020 DOI: 10.3892/ijo.2020.5122 Abstract. Epigenetic modifications are important contributors 1. Introduction to the regulation of genes within the chromatin. The poly- comb repressive complex 2 (PRC2) is a multi‑subunit protein Epigenetic modifications, including DNA methylation complex that is involved in silencing gene expression through and histone modifications, play an important role in gene the trimethylation of lysine 27 at histone 3 (H3K27me3). The regulation. The dysregulation of these modifications can dysregulation of this modification has been associated with result in pathogenicity, including tumorigenicity. Research tumorigenicity through the increased repression of tumour has indicated an important influence of the trimethylation suppressor genes via condensing DNA to reduce access to the modification at lysine 27 on histone H3 (H3K27me3) within transcription start site (TSS) within tumor suppressor gene chromatin. This methylation is involved in the repression promoters. In the present review, the core proteins of PRC2, as of multiple genes within the genome by condensing DNA well as key accessory proteins, will be described. In addition, to reduce access to the transcription start site (TSS) within mechanisms controlling the recruitment of the PRC2 complex gene promoter sequences (1). The recruitment of H1.2, an H1 to H3K27 will be outlined. Finally, literature identifying the histone subtype, by the H3K27me3 modification has been a role of PRC2 in breast cancer proliferation, apoptosis and suggested as a mechanism for mediating this compaction (1). -

Ablation of Ezh2 in Neural Crest Cells Leads to Hirschsprung's Disease-Like Phenotype in Mice
bioRxiv preprint doi: https://doi.org/10.1101/265868; this version posted February 15, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Ablation of Ezh2 in neural crest cells leads to Hirschsprung’s disease-like phenotype in mice Hana Kim, Ingeborg M. Langohr, Mohammad Faisal, Margaret McNulty, Caitlin Thorn, Joomyeong Kim* Department of Biological Sciences, Louisiana State University, Baton Rouge, LA 70803, USA. Correspondence should be forwarded to: [email protected], 225-578-7692(ph), or 225-578-2597(fax) Running title: Function of Ezh2 in enteric neural crest cell development 1 bioRxiv preprint doi: https://doi.org/10.1101/265868; this version posted February 15, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract In the current study, we examined the role of Ezh2 as an epigenetic modifier for the enteric neural crest cell development through H3K27me3. Ezh2 conditional null mice were viable up to birth, but died within the first hour of life. In addition to craniofacial defects, Ezh2 conditional null mice displayed reduced number of ganglion cells in the enteric nervous system. RT-PCR and ChIP assays indicated aberrant up-regulation of Zic1, Pax3, and Sox10 and loss of H3K27me3 marks in the promoter regions of these genes in the myenteric plexus. Overall, these results suggest that Ezh2 is an important epigenetic modifier for the enteric neural crest cell development through repression of Zic1, Pax3, and Sox10.