The Dynamic Fungal Cell
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

IB DIPLOMA PROGRAMME Debora M
OXFORD IB PREPARED BIOLOGY IB DIPLOMA PROGRAMME Debora M. Primrose Contents Introduction iv 9 Plant biology (AHL) 1 Cell biology 9.1 Transport in the xylem of plants 103 9.2 Transport in the phloem of plants 107 1.1 Introduction to cells 2 9.3 Growth in plants 110 1.2 Ultrastructure of cells 4 9.4 Reproduction in plants 113 1.3 Membrane structure 6 1.4 Membrane transport 7 10 Genetics and evolution (AHL) 1.5 The origin of cells 9 10.1 Meiosis 117 1.6 Cell division 11 10.2 Inheritance 121 2 Molecular biology 10.3 Gene pools and speciation 125 2.1 Molecules to metabolism 14 11 Animal physiology (AHL) 2.2 Water 15 11.1 Antibody production and vaccination 128 2.3 Carbohydrates and lipids 16 11.2 Movement 133 2.4 Proteins 20 11.3 The kidney and osmoregulation 137 2.5 Enzymes 21 11.4 Sexual reproduction 141 2.6 Structure of DNA and RNA 23 2.7 DNA replication, transcription and translation 24 12 Data-based and practical questions 147 2.8 Cell respiration 26 2.9 Photosynthesis 28 A Neurobiology and behaviour 3 Genetics A.1 Neural development 157 A.2 The human brain 159 3.1 Genes 30 A.3 Perception of stimuli 161 3.2 Chromosomes 32 A.4 Innate and learned behaviour (AHL) 165 3.3 Meiosis 33 A.5 Neuropharmacology (AHL) 167 3.4 Inheritance 35 A.6 Ethology (AHL) 169 3.5 Genetic modification and biotechnology 37 B Biotechnology and bioinformatics 4 Ecology B.1 Microbiology: organisms in industry 172 4.1 Species, communities and ecosystems 40 B.2 Biotechnology in agriculture 174 4.2 Energy flow 43 B.3 Environmental protection 178 4.3 Carbon cycling 45 -

The Still Valid Fluid Mosaic Model for Molecular Organization of Biomembranes: Accumulating Data Confirm It
DISCOVERIES 2013, Oct-Dec; 1(1): e7 DOI: 10.15190/d.2013.7 The still actual fluid mosaic model for membrane organization Focused REVIEW The still valid fluid mosaic model for molecular organization of biomembranes: accumulating data confirm it Mircea Leabu1,* 1University of Medicine and Pharmacy “Carol Davila”, Department of Cellular and Molecular Medicine; “Victor Babes” National Institute of Pathology; University of Bucharest, Research Center for Applied Ethics *Correspondence to: Mircea Leabu, PhD, “Victor Babes” National Institute of Pathology, 99101, Splaiul Independentei, 050096, Bucharest, Romania; E-mail: [email protected] Citation: Leabu M. The still valid fluid mosaic model for molecular organization of biomembranes: accumulating data confirm it. Discoveries 2013, Oct-Dec; 1(1): e7. DOI: 10.15190/d.2013.7 ABSTRACT Keywords: membrane organization, membrane fluidity, membrane microdomains, Singer-Nicolson More than forty years passed since Singer and model, science history Nicolson launched the fluid mosaic model related to molecular organization and dynamics of cell Introduction membranes, applicable to endomembranes as well. For 20 years I and perhaps many other professors During this period of time, that will reach half a have been teaching students in medicine about the century soon, accumulating data all confirm, but not molecular organization of the cell membrane, infirm the brilliant idea of such a model. presenting with a high enthusiasm the fluid mosaic Sometimes, the results developed the model in a model launched by Seymour Jonathan Singer and very impacting manner, as was the case with the Garth L. Nicolson in 19721, and considering it as an introduction of the membrane microdomain concept inspired, still actual scientific and even pedagogic (mainly lipid rafts organization). -

The Structure and Function of the Plasma Membrane
120 4 The Structure and Function of the Plasma Membrane 4.1 An Overview of Membrane Functions The outer walls of a house or car provide a strong, inflexible 4.2 A Brief History of Studies on Plasma Membrane barrier that protects its human inhabitants from an unpredictable Structure and harsh external world. You might expect the outer boundary of a living cell to be constructed of an equally tough and impenetra- 4.3 The Chemical Composition of Membranes ble barrier because it must also protect its delicate internal con- 4.4 The Structure and Functions of Membrane Proteins tents from a nonliving, and often inhospitable, environment. Yet 4.5 Membrane Lipids and Membrane Fluidity cells are separated from the external world by a thin, fragile 4.6 The Dynamic Nature of the Plasma Membrane structure called the plasma membrane that is only 5 to 10 nm wide. It would require about five thousand plasma membranes 4.7 The Movement of Substances Across Cell stacked one on top of the other to equal the thickness of a Membranes single page of this book. 4.8 Membrane Potentials and Nerve Impulses Because it is so thin, no hint of the plasma membrane is THE HUMAN PERSPECTIVE: Defects in Ion Channels and detected when a section of a cell is examined under a light Transporters as a Cause of Inherited Disease microscope. In fact, it wasn’t until the late 1950s that techniques for preparing and staining tissue had progressed to the point EXPERIMENTAL PATHWAYS: The Acetylcholine Receptor where the plasma membrane could be resolved in the electron Three-dimensional, X-ray crystallographic structure of a 2-adrenergic receptor ( 2-AR), which is a member of the G protein-coupled receptor (GPCR) superfamily. -

The Cell Membrane Surrounds All Living Cells and Is the Most
The Cell membrane surrounds all living cells and is the most important organelle, there is also a similar plasma membrane that surrounds all the organelles except for the ribosome. The membrane controls how and what substances can move in and out of the cell/organelle The structure of the membrane is often referred to as the “Fluid Mosaic Model”; this is because of the way it is structured It is composed of phospholipids, proteins, and carbohydrates, which are arranged in a fluid mosaic structure. The phospholipids are arranged in a “bilayer”. With their hydrophilic (water attracting) phosphate heads facing outwards and their hydrophobic (water fearing) tails facing in towards the middle of the bilayer. The hydrophobic layer acts as a barrier to all but the smallest molecules and effectively isolating the two sides of the membranes. Some membranes contain phospholipids with different fatty acids, which affect the strength and flexibility. Animal cells also have cholesterol linking the fatty acids together and so stabilising and strengthening then membrane The proteins usually span from one side of the bilayer to the other. These are called integral proteins. But some sit on one side of the bilayer, these are called peripheral proteins. Proteins comprise approximately 50% of the mass of the membrane. The integral proteins (ones which span across the whole bilayer) are usually involved in the transporting of substances across the membrane. The proteins that are on the inside of the bilayer are often attached to the cytoskeleton and are involved in maintaining the cell’s shape. They may also be enzymes for catalysing reactions. -

2 the Structure and Ultrastructure of the Cell Gunther Neuhaus Institut Fu¨R Zellbiologie, Freiburg, Germany
2 The Structure and Ultrastructure of the Cell Gunther Neuhaus Institut fu¨r Zellbiologie, Freiburg, Germany 2.1 Cell Biology . .........................40 2.2.7.6 Isolating Secondary Walls . 107 2.1.1 Light Microscopy . 43 2.2.8 Mitochondria . 109 2.1.2 Electron Microscopy . 45 2.2.8.1 Shape Dynamics and Reproduction . 110 2.2.8.2 Membranes and Compartmentalization in 2.2 The Plant Cell . .........................46 Mitochondria . 112 2.2.1 Overview . 46 2.2.9 Plastids . 113 2.2.2 The Cytoplasm . 50 2.2.9.1 Form and Ultrastructure of 2.2.2.1 The Cytoskeleton . 51 Chloroplasts . 114 2.2.2.2 Motor Proteins and Cellular Kinetic 2.2.9.2 Other Plastid Types, Starches . 116 Processes . 55 2.2.2.3 Flagella and Centrioles . 57 2.3 Cell Structure in Prokaryotes ............. 119 2.2.3 The Cell Nucleus . 59 2.3.1 Reproduction and Genetic Apparatus . 122 2.2.3.1 Chromatin . 60 2.3.2 Bacterial Flagella . 124 2.2.3.2 Chromosomes and Karyotype . 63 2.3.3 Wall Structures . 125 2.2.3.3 Nucleoli and Preribosomes . 64 2.2.3.4 Nuclear Matrix and Nuclear Membrane . 65 2.4 The Endosymbiotic Theory and the 2.2.3.5 Mitosis and the Cell Cycle . 66 Hydrogen Hypothesis . ............. 125 2.2.3.6 Cell Division . 73 2.4.1 Endocytobiosis . 126 2.2.3.7 Meiosis . 73 2.4.2 Origin of Plastids and Mitochondria by 2.2.3.8 Crossing-Over . 79 Symbiogenesis . 127 2.2.3.9 Syngamy . 79 2.2.4 Ribosomes . -
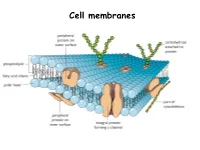
Applied Science Cell Membrane
Cell membranes Plasma Membrane Structure and Function The plasma membrane separates the internal environment of the cell from its surroundings. The plasma membrane is a phospholipid bilayer with embedded proteins. The plasma membrane has a fluid consistency and a mosaic pattern of embedded proteins. The cell-surface membrane controls the movement of substances into and out of the cell. Cell membranes are made up of…. Phospholipids Phospholipids allow…. • Lipid- soluble substances to enter and leave the cell • Prevent water-soluble substances entering and leaving the cell • Make the membrane flexible and self-sealing. Make a note of this information Proteins are also found in the bilayer…. http://www.youtube.com/watch?v=moPJkCbKjBs&feat ure=rhttp://www.youtube.com/watch?v=moPJkCbKj Bs&feature=relatedelated http://www.youtube.com/watch?v=moPJkCbKjBs&f eature=related The fluid-mosaic model of the cell-surface membrane http://highered.mcgraw- hill.com/sites/0072495855/student_view0/chapter 2/animation__how_facilitated_diffusion_works.ht ml Movement across membranes Molecules can pass across membranes by: • Simple diffusion • Facilitated diffusion • Active transport The Permeability of the Plasma Membrane The plasma membrane is partially permeable. Macromolecules cannot pass through because of their size, and tiny charged molecules do not pass through the nonpolar interior of the membrane. Small, uncharged molecules such as oxygen and carbon dioxide pass through the membrane, down their concentration gradient. How molecules cross the plasma membrane Diffusion Diffusion is the passive movement of molecules from a higher to a lower concentration until equilibrium is reached. Gases move through plasma membranes by diffusion. Process of diffusion Gas exchange in the lungs occurs by diffusion Facilitated diffusion During facilitated diffusion, substances pass through a carrier protein following their concentration gradients. -

Structure and Function of Cell Membranes1,2 Marvin H
STRUCTURE AND FUNCTION OF CELL MEMBRANES1,2 MARVIN H. STROMER AND WILLIAM J. REVILLE Iowa State University3, Ames, Iowa 50010 INTRODUCTION The purpose of this presentation is to provide some backgrmnd information regarding the organization and function of cell membranes, in general, and then to focus attention on the membranes of the skeletal muscle cell. The interrelationships of the specialized membranes of the skeletal muscle cell will be discussed in order to develop a framework into which the succeeding three papers can be integrated. In attempting to ascribe a purpose or function to any biological membrane in a cell, one is rapidly forced to conclude that membranes sequester or segregate components of the cell or the cell itself from its environment and regulate what external stimuli or components may enter the cell. This is the situation for a wide variety of cells, including muscle. Mernbranes inside a cell may maintain favorable microenvironments within closed tubular or vesicular systems and prmide a structural matrix on which enzymes may be favorably oriented to interact with substrates and on which certain essential features such as ribosomes may be located to maximize their efficiency. Processes such as protein synthesized for export may be regulated or modulated by interactions with membranes. MEMBRANE MODELS a. Unit membrane model Research on membranes from a variety of sources is a very active and diverse area of investigation. The two models that attempt to explain membrane structure and components are the unit membrane or bimolecular leaflet model of Danielli-Davson-Robertson (6, 2) and the fluid mosaic model of Singer and Nicolson (28). -

Endoplasmic Reticulum Layers of Endoplasmic Reticulum with Attached Ribosomes
TheThe CytoplasmCytoplasm Li Shulei [email protected] DepartmentDepartment ofof HistologyHistology && EmbryologyEmbryology CellCell componentscomponents CytoplasmCytoplasm PlasmaPlasma membranemembrane OrganellesOrganelles CytoplasmicCytoplasmic depositsdeposits CytoskeletonCytoskeleton CytosolCytosol (( MatrixMatrix )) NucleusNucleus PlasmaPlasma membranemembrane StructureStructure Thickness:7.5nm~10nmThickness:7.5nm~10nm UnitUnit membranemembrane FluidFluid mosaicmosaic modelmodel MainMain functionsfunctions TransmemebraneTransmemebrane transporttransport SignalSignal transmembranetransmembrane transductiontransduction PlasmaPlasma membranemembrane trilaminar appearance PlasmaPlasma membranemembrane Hydrophilic portion The ultrastructure and molecular organization of the cell membrane. The dark lines at left represent the two dense layers observed in the electron microscope. Cholesterol breaks up the close packing of phospholipid chains, and makes the membrane more fluid. The lipid composition of each half of the bilayer is different. PlasmaPlasma membranemembrane phospholipid integral double layer protein peripheraltransmembran proteineproteins A: The fluid mosaic model of membrane structure. B: Membrane cleavage occurs when cell is frozen and fractured into two parts along the hydrophobic interactions . PlasmaPlasma membranemembrane molecular structure of the plasma membrane. one-pass and multipass transmembrane proteins, peripheral protein proteins are present mainly in the cytoplasmic face. (1)(1) MitochondriaMitochondria -

7.2 Cell Structure Lesson Overview Cell Structure
7.2 cell structure Lesson Overview Cell Structure Cell Organization The eukaryotic cell has two major parts: the nucleus and the cytoplasm. cytoplasm - fluid portion of the cell outside the nucleus. -Prokaryotic cells have cytoplasm too. Eukaryotic cells contain many organelles - membrane bound structures that perform specialized tasks. Lesson Overview Cell Structure The Nucleus nucleus - control center of the cell. - contains the cell’s DNA - surrounded by a nuclear envelope made of 2 membranes. - contains nucleolus = site of ribosome synthesis Lesson Overview Cell Structure The Nucleus The nuclear envelope contains nuclear pores, which allow material to move into and out of the nucleus. Proteins, RNA, and other molecules move through the nuclear pores to and from the rest of the cell Lesson Overview Cell Structure The Nucleus DNA in the nucleus is usually seen as Chromatin - long, thin strings of DNA. When a cell divides, its chromatin condenses into shorter, thicker chromosomes. Chromosomes contain the genetic information (DNA) passed to the offspring. Lesson Overview Cell Structure Vacuoles and Vesicles cells contain large, saclike, membrane-enclosed structures called vacuoles that store materials like water, salts, proteins, and carbohydrates. Lesson Overview Cell Structure Vacuoles and Vesicles Plant cells have a single, large central vacuole. Turgid pressure of the central vacuole increases their rigidity helping maintain plant structure. Lesson Overview Cell Structure Vacuoles and Vesicles Vacuoles are present in some unicellular organisms and in some animals. Lesson Overview Cell Structure Vacuoles and Vesicles Many eukaryotic cells contain smaller membrane-enclosed structures called vesicles. Vesicles store and move materials between organelles and to and from the cell surface. -

Cell – Structure and Function MODULE - 1 Diversity and Evolution of Life
Cell – Structure and Function MODULE - 1 Diversity and Evolution of Life 4 Notes CELL – STRUCTURE AND FUNCTION INTRODUCTION All organisms are composed of structural and functional units of life called ‘cells’. The body of some organisms like bacteria, protozoans and some algae is made up of a single cell whereas the body of higher fungi, plants and animals are composed of many cells. Human body is built of about one trillion cells. Cells vary in size and structure as they are specialized to perform different functions. But the basic components of the cell are common to all biological cells. This lesson deals with the structure common to all types of the cells. You will also learn about the kinds of cell division and the processes involved therein in this lesson. OBJECTIVES After completing this lesson, you will be able to : z justify that cell is the basic structural and functional unit of all organisms; z list the components of the cell and state cell theory; z differentiate between prokaryotic and eukaryotic cells; z differentiate between plant and animal cells; z illustrate the structure of plant and animal cells by drawing labelled diagrams; z describe the structure and functions of plasma membrane, cell wall, endoplasmic reticulum (ER), cilia, flagella, nucleus, ribosomes, mitochondria, chloroplasts, golgi body, peroxisome, glyoxysome and lysosome; z describe the general importance of the cell molecules-water, mineral ions, carbohydrates, lipids, amino acids, proteins, nucleotides, nucleic acids, enzymes, vitamins, hormones, steroids and alkaloids; z justify the need for cell division; z describe various phases of cell cycle; z explain the term karyotype and mention the karyotype analysis and its significance. -

Chapter 03 Lecture
Chapter 03 Lecture 1 The Prokaryotic Cell Pilus Ribosomes Cytoplasm Chromosome (DNA) Nucleoid Cell wall Flagellum (b) 0.5 µm Capsule Cell wall Cytoplasmic membrane (a) The Prokaryotic Cell 3.4. The Cytoplasmic Membrane . Cytoplasmic membrane defines boundary of cell • Phospholipid bilayer embedded with proteins • Hydrophobic tails face in; hydrophilic tails face out • Serves as semipermeable membrane • Proteins serve numerous functions • Selective gates • Sensors of environmental conditions Phospholipid bilayer • Fluid mosaic model: proteins drift about in lipid bilayer Hydrophilic head Hydrophobic tail Proteins 3.4. The Cytoplasmic Membrane . Cytoplasmic membrane defines boundary of cell (continued…) • Bacteria and Archaea have same general structure of cytoplasmic membranes • Distinctly different lipid compositions • Lipid tails of Archaea are not fatty acids and are connected differently to glycerol Permeability of Lipid Bilayer . Cytoplasmic membrane is selectively permeable • O2, CO2, N2, small hydrophobic molecules, and water pass freely • Some cells facilitate water passage with aquaporins • Other molecules must be moved across membrane via transport systems Pass through easily: Passes through: Do not pass through: Gases (O2, CO2, N2) Water Sugars Small hydrophobic Ions molecules Amino acids ATP Water Macromolecules Aquaporin (a) The cytoplasmic membrane is selectively permeable. Gases, small (b) Aquaporins allow water to pass through the cytoplasmic membrane hydrophobic molecules, and water are the only substances that more easily. -

MEMBRANE MODEL: the Bubble Lab
MEMBRANE MODEL: The Bubble Lab The cell’s plasma membrane is a phospholipid bilayer with protein molecules imbedded in it. The protein molecules transport other molecules through the membrane and into or out of the cell. All of the membranes in the cell (nuclear envelop, endoplasmic reticulum, membranes in the chloroplasts and mitochondria) are essentially the same as the plasma membrane. Soap bubbles are bilayers very similar to phospholipids membranes, so they can be used to investigate some of the properties of the cell membrane. Procedure: 1) Immerse the membrane holder into the pan of soap solution. Raise it out of the pan and allow the excess soap to drip off. Hold up the membrane holder and demonstrate the following characteristics of a lipid bilayer. • Fluidity: The cell membrane is called the Fluid Mosaic Model. This means that the membrane is made of a pattern of many small molecules that are moving around and shifting position. 2) Observe the light shining of the surface of the soap film? Notice the movement in the light pattern, demonstrating that the molecules of the film are constantly in motion. • Flexibility: A lipid bilayer is a fluid arrangement in which the molecules move freely through the plane of the bilayer- reorganizing into almost any shape. 3) Twist the two straw handles in opposite directions and bend the film into different configurations. Note what happens to the soap film. The soap bilayer is actually less flexible than a cell membrane because a cell membrane is supported on both sides, one side by the cytoplasm and the other tissue fluids, or other cells.