The Temporal Mechanisms Guiding Interneuron Differentiation in the Spinal Cord
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Neural Control of Movement: Motor Neuron Subtypes, Proprioception and Recurrent Inhibition
List of Papers This thesis is based on the following papers, which are referred to in the text by their Roman numerals. I Enjin A, Rabe N, Nakanishi ST, Vallstedt A, Gezelius H, Mem- ic F, Lind M, Hjalt T, Tourtellotte WG, Bruder C, Eichele G, Whelan PJ, Kullander K (2010) Identification of novel spinal cholinergic genetic subtypes disclose Chodl and Pitx2 as mark- ers for fast motor neurons and partition cells. J Comp Neurol 518:2284-2304. II Wootz H, Enjin A, Wallen-Mackenzie Å, Lindholm D, Kul- lander K (2010) Reduced VGLUT2 expression increases motor neuron viability in Sod1G93A mice. Neurobiol Dis 37:58-66 III Enjin A, Leao KE, Mikulovic S, Le Merre P, Tourtellotte WG, Kullander K. 5-ht1d marks gamma motor neurons and regulates development of sensorimotor connections Manuscript IV Enjin A, Leao KE, Eriksson A, Larhammar M, Gezelius H, Lamotte d’Incamps B, Nagaraja C, Kullander K. Development of spinal motor circuits in the absence of VIAAT-mediated Renshaw cell signaling Manuscript Reprints were made with permission from the respective publishers. Cover illustration Carousel by Sasha Svensson Contents Introduction.....................................................................................................9 Background...................................................................................................11 Neural control of movement.....................................................................11 The motor neuron.....................................................................................12 Organization -

Innovations Present in the Primate Interneuron Repertoire
Article Innovations present in the primate interneuron repertoire https://doi.org/10.1038/s41586-020-2781-z Fenna M. Krienen1,2 ✉, Melissa Goldman1,2, Qiangge Zhang2,3, Ricardo C. H. del Rosario2, Marta Florio1,2, Robert Machold4, Arpiar Saunders1,2, Kirsten Levandowski2,3, Heather Zaniewski2,3, Received: 19 July 2019 Benjamin Schuman4, Carolyn Wu3, Alyssa Lutservitz1,2, Christopher D. Mullally1,2, Nora Reed1,2, Accepted: 1 July 2020 Elizabeth Bien1,2, Laura Bortolin1,2, Marian Fernandez-Otero2,5, Jessica D. Lin2, Alec Wysoker2, James Nemesh2, David Kulp2, Monika Burns5, Victor Tkachev6,7,8, Richard Smith9,10, Published online: xx xx xxxx Christopher A. Walsh9,10, Jordane Dimidschstein2, Bernardo Rudy4,11, Leslie S. Kean6,7,8, Check for updates Sabina Berretta5,12,13, Gord Fishell2,14, Guoping Feng2,3 & Steven A. McCarroll1,2 ✉ Primates and rodents, which descended from a common ancestor around 90 million years ago1, exhibit profound diferences in behaviour and cognitive capacity; the cellular basis for these diferences is unknown. Here we use single-nucleus RNA sequencing to profle RNA expression in 188,776 individual interneurons across homologous brain regions from three primates (human, macaque and marmoset), a rodent (mouse) and a weasel (ferret). Homologous interneuron types—which were readily identifed by their RNA-expression patterns—varied in abundance and RNA expression among ferrets, mice and primates, but varied less among primates. Only a modest fraction of the genes identifed as ‘markers’ of specifc interneuron subtypes in any one species had this property in another species. In the primate neocortex, dozens of genes showed spatial expression gradients among interneurons of the same type, which suggests that regional variation in cortical contexts shapes the RNA expression patterns of adult neocortical interneurons. -

Spinal Cord Organization
Lecture 4 Spinal Cord Organization The spinal cord . Afferent tract • connects with spinal nerves, through afferent BRAIN neuron & efferent axons in spinal roots; reflex receptor interneuron • communicates with the brain, by means of cell ascending and descending pathways that body form tracts in spinal white matter; and white matter muscle • gives rise to spinal reflexes, pre-determined gray matter Efferent neuron by interneuronal circuits. Spinal Cord Section Gross anatomy of the spinal cord: The spinal cord is a cylinder of CNS. The spinal cord exhibits subtle cervical and lumbar (lumbosacral) enlargements produced by extra neurons in segments that innervate limbs. The region of spinal cord caudal to the lumbar enlargement is conus medullaris. Caudal to this, a terminal filament of (nonfunctional) glial tissue extends into the tail. terminal filament lumbar enlargement conus medullaris cervical enlargement A spinal cord segment = a portion of spinal cord that spinal ganglion gives rise to a pair (right & left) of spinal nerves. Each spinal dorsal nerve is attached to the spinal cord by means of dorsal and spinal ventral roots composed of rootlets. Spinal segments, spinal root (rootlets) nerve roots, and spinal nerves are all identified numerically by th region, e.g., 6 cervical (C6) spinal segment. ventral Sacral and caudal spinal roots (surrounding the conus root medullaris and terminal filament and streaming caudally to (rootlets) reach corresponding intervertebral foramina) collectively constitute the cauda equina. Both the spinal cord (CNS) and spinal roots (PNS) are enveloped by meninges within the vertebral canal. Spinal nerves (which are formed in intervertebral foramina) are covered by connective tissue (epineurium, perineurium, & endoneurium) rather than meninges. -
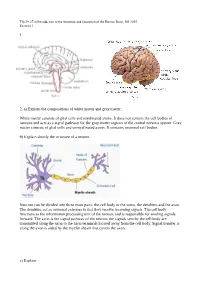
1. 2. A) Explain the Compositions of White Matter and Gray
Tfy-99.2710 Introduction to the Structure and Operation of the Human Brain, fall 2015 Exercise 1 1. 2. a) Explain the compositions of white matter and gray matter. White matter consists of glial cells and myelinated axons. It does not contain the cell bodies of neurons and acts as a signal pathway for the gray matter regions of the central nervous system. Gray matter consists of glial cells and unmyelinated axons. It contains neuronal cell bodies. b) Explain shortly the structure of a neuron. Neurons can be divided into three main parts: the cell body or the soma, the dendrites and the axon. The dendrites act as neuronal antennas in that they receive incoming signals. The cell body functions as the information processing unit of the neuron, and is responsible for sending signals forward. The axon is the signal pathway of the neuron; the signals sent by the cell body are transmitted along the axon to the axon terminals located away from the cell body. Signal transfer is along the axon is aided by the myelin sheath that covers the axon. c) Explain: Tfy-99.2710 Introduction to the Structure and Operation of the Human Brain, fall 2015 Exercise 1 i) myelin Membrane that wraps around axons. Formed from glial support cells: oligodendrogolia in the central nervous system and Schwann cells in the peripheral nervous system. Myelin facilitates signal transfer along axons by allowing action potentials to skip between the nodes of Ranvier (saltatory conduction). ii) receptor Molecule specialized in receiving a chemical signal by binding a specific neurotransmitter. -

Cortex Brainstem Spinal Cord Thalamus Cerebellum Basal Ganglia
Harvard-MIT Division of Health Sciences and Technology HST.131: Introduction to Neuroscience Course Director: Dr. David Corey Motor Systems I 1 Emad Eskandar, MD Motor Systems I - Muscles & Spinal Cord Introduction Normal motor function requires the coordination of multiple inter-elated areas of the CNS. Understanding the contributions of these areas to generating movements and the disturbances that arise from their pathology are important challenges for the clinician and the scientist. Despite the importance of diseases that cause disorders of movement, the precise function of many of these areas is not completely clear. The main constituents of the motor system are the cortex, basal ganglia, cerebellum, brainstem, and spinal cord. Cortex Basal Ganglia Cerebellum Thalamus Brainstem Spinal Cord In very broad terms, cortical motor areas initiate voluntary movements. The cortex projects to the spinal cord directly, through the corticospinal tract - also known as the pyramidal tract, or indirectly through relay areas in the brain stem. The cortical output is modified by two parallel but separate re entrant side loops. One loop involves the basal ganglia while the other loop involves the cerebellum. The final outputs for the entire system are the alpha motor neurons of the spinal cord, also called the Lower Motor Neurons. Cortex: Planning and initiation of voluntary movements and integration of inputs from other brain areas. Basal Ganglia: Enforcement of desired movements and suppression of undesired movements. Cerebellum: Timing and precision of fine movements, adjusting ongoing movements, motor learning of skilled tasks Brain Stem: Control of balance and posture, coordination of head, neck and eye movements, motor outflow of cranial nerves Spinal Cord: Spontaneous reflexes, rhythmic movements, motor outflow to body. -

Intersegmental Interneurons Can Control the Gain of Reflexes in Adjacent Segments of the Locust by Their Action on Nonspiking Local Interneurons
The Journal of Neuroscience, September 1989, g(9): 30303039 Intersegmental Interneurons Can Control the Gain of Reflexes in Adjacent Segments of the Locust by Their Action on Nonspiking Local Interneurons Gilles Laurent and Malcolm Burrows Department of Zoology, University of Cambridge, Cambridge CB2 3EJ, England The gain of local reflexes of one leg of a locust can be altered partmentalized, with synaptic inputs and their associated by mechanosensory inputs generated by movements of or conductance changes restricted to particular branches. In tactile inputs to an adjacent leg. Touching the mesothoracic this way, an individual nonspiking neuron could contribute tarsus, for example, increases the number of spikes that are simultaneously to several local circuits. The inputs from dif- produced by the metathoracic slow extensor tibiae motor ferent intersegmental interneurons could then modulate these neuron and enhances the depolarization of flexor tibiae mo- pathways independently. tor neuron in response to imposed movements of the chor- dotonal organ in the ipsilateral hind femur. The sensory in- Nonspiking interneurons in the metathoracic ganglion of the formation from the middle leg is conveyed directly to locust receive direct inputs from mechanosensory afferents on nonspiking interneurons and motor neurons controlling the one hindleg and are essential elements in local reflex movements movements of the hindleg by a population of mesothoracic of that leg (Laurent and Burrows, 1988; Burrows et al., 1988). intersegmental interneurons (Laurent and Burrows, 1989). They also receive direct inputs from intersegmental intemeu- The metathoracic nonspiking interneurons receive direct in- rons that process the mechanosensory inputs from an ipsilateral puts from receptors on a hindleg and are, therefore, a point middle leg (Laurent and Burrows, 1989). -

Spinal Reflexes
Spinal Reflexes Lu Chen, Ph.D. MCB, UC Berkeley 1 Simple reflexes such as stretch reflex require coordinated contraction and relaxation of different muscle groups Categories of Muscle Based on Direction of Motion Flexors Æ reduce the angle of joints Extensors Æ increase the angle of joints Categories of Muscle Based on Movement Agonist Æmuscle that serves to move the joint in the same direction as the studied muscle Antagonist Æ muscle that moves the joint in the opposite direction 2 1 Muscle Spindles •Small encapsulated sensory receptors that have a Intrafusal muscle spindle-like shape and are located within the fibers fleshy part of the muscle •In parallel with the muscle fibers capsule •Does not contribute to the overall contractile Sensory force endings •Mechanoreceptors are activated by stretch of the central region Afferent axons •Due to stretch of the whole muscle Efferent axons (including intrafusal f.) •Due to contraction of the polar regions of Gamma motor the intrafusal fibers endings 3 Muscle Spindles Organization 2 kinds of intrafusal muscle fibers •Nuclear bag fibers (2-3) •Dynamic •Static •Nuclear chain fibers (~5) •Static 2 types of sensory fibers •Ia (primary) - central region of all intrafusal fibers •II (secondary) - adjacent to the central region of static nuclear bag fibers and nuclear chain fibers Intrafusal fibers stretched Sensory ending stretched, (loading the spindle) increase firing Muscle fibers lengthens Sensory ending stretched, (stretched) increase firing Spindle unloaded, Muscle fiber shortens decrease firing 4 2 Muscle Spindles Organization Gamma motor neurons innervate the intrafusal muscle fibers. Activation of Shortening of the polar regions gamma neurons of the intrafusal fibers Stretches the noncontractile Increase firing of the center regions sensory endings Therefore, the gamma motor neurons provide a mechanism for adjusting the sensitivity of the muscle spindles. -

Retinal Inputs Signal Astrocytes to Recruit Interneurons Into Visual Thalamus
Retinal inputs signal astrocytes to recruit interneurons into visual thalamus Jianmin Sua,1, Naomi E. Charalambakisb,1, Ubadah Sabbagha,c, Rachana D. Somaiyaa,c, Aboozar Monavarfeshania,d, William Guidob,2, and Michael A. Foxa,d,e,2 aCenter for Neurobiology Research, Fralin Biomedical Research Institute at Virginia Tech Carilion, Roanoke, VA 24016; bDepartment of Anatomical Sciences and Neurobiology, University of Louisville School of Medicine, Louisville, KY 40202; cGraduate Program in Translational Biology, Medicine, and Health, Virginia Tech, Blacksburg, VA 24061; dDepartment of Biological Sciences, Virginia Tech, Blacksburg, VA 24061; and eDepartment of Pediatrics, Virginia Tech Carilion School of Medicine, Roanoke, VA 24016 Edited by Carol Ann Mason, Columbia University, New York, NY, and approved December 24, 2019 (received for review July 29, 2019) Inhibitory interneurons comprise a fraction of the total neurons in collection of cell types based on their distribution, gene and the visual thalamus but are essential for sharpening receptive field neurochemical expression, and projection patterns (5–7, 25). properties and improving contrast-gain of retinogeniculate trans- Despite the presence of GABAergic interneurons in the visual mission. During early development, these interneurons undergo thalamus of the adult mouse, these cells are largely absent at long-range migration from germinal zones, a process regulated by birth when RGC axons innervate these nuclei (12). Also lacking the innervation of the visual thalamus by retinal ganglion cells. from the visual thalamus at these early ages are nonretinal inputs Here, using transcriptomic approaches, we identified a motogenic that arise from the neocortex, brainstem, and the thalamic re- cue, fibroblast growth factor 15 (FGF15), whose expression in the ticular nucleus. -

Impaired Interneuron Development in a Novel Model of Neonatal Brain Injury
New Research Development Impaired Interneuron Development in a Novel Model of Neonatal Brain Injury Helene Lacaille,1 Claire-Marie Vacher,1 Dana Bakalar,1 Jiaqi J. O’Reilly,1,2 Jacquelyn Salzbank,1,2 and Anna A. Penn1,2,3 https://doi.org/10.1523/ENEURO.0300-18.2019 1Center for Neuroscience, Children’s National Health System, Washington, DC 20010, 2Institute for Biomedical Sciences, George Washington University, Washington, DC 20037, and 3Fetal Medicine Institute, Neonatology, Children’s National Health System, Washington, DC 20010 Abstract Prematurity is associated with significantly increased risk of neurobehavioral pathologies, including autism and schizophrenia. A common feature of these psychiatric disorders is prefrontal cortex (PFC) inhibitory circuit disruption due to GABAergic interneuron alteration. Cortical interneurons are generated and migrate throughout late gestation and early infancy, making them highly susceptible to perinatal insults such as preterm birth. Term and preterm PFC pathology specimens were assessed using immunohistochemical markers for interneurons. Based on the changes seen, a new preterm encephalopathy mouse model was developed to produce similar PFC interneuron loss. Maternal immune activation (MIA; modeling chorioam- nionitis, associated with 85% of extremely preterm births) was combined with chronic sublethal hypoxia (CSH; modeling preterm respiratory failure), with offspring of both sexes assessed anatomically, molecularly and neurobehaviorally. In the PFC examined from the human preterm samples compared to matched term samples at corrected age, a decrease in somatostatin (SST) and calbindin (CLB) interneurons was seen in upper cortical layers. This pattern of interneuron loss in upper cortical layers was mimicked in the mouse PFC following the combination of MIA and CSH, but not after either insult alone. -

The Transcriptional Landscape of Cortical Interneurons Underlies In
bioRxiv preprint doi: https://doi.org/10.1101/481036; this version posted November 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 The transcriptional landscape of cortical interneurons underlies in-vivo brain function and schizophrenia 2 risk 3 4 Kevin M Anderson1, Meghan A Collins1, Rowena Chin1, Tian Ge2,3, Monica D Rosenberg1, Avram J 5 Holmes1,3,4* 6 7 1Department of Psychology, Yale University, New Haven, Connecticut 06520 8 2Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts 9 General Hospital, Boston, MA 02114, USA 10 3Department of Psychiatry, Massachusetts General Hospital, Harvard Medical School, 11 Boston, MA 02114, USA 4 12 Department of Psychiatry, Yale University, New Haven, Connecticut 06520, USA 13 *Corresponding author. 14 15 Author contributions. KMA and AJH designed the research. KMA conducted the research. MAC, RC, 16 MDR, TG provided analytic support. KMA and AJH wrote the manuscript and made figures. All authors 17 edited the manuscript. 18 19 Acknowledgements: This work was supported by the National Institute of Mental Health (Grant 20 K01MH099232 to A.J.H.), the National Science Foundation (DGE-1122492 to K.M.A.), and the National 21 Institute on Aging (K99AG054573 to T.G.). Analyses were made possible by the high-performance 22 computing facilities provided through the Yale Center for Research Computing. We thank B.J. Casey, 23 Danielle Gerhard, Lauren Patrick, and Erica Ho for their feedback on early versions of the project. -
Nervous System - Neurons
Nervous System - Neurons Biol 105 Chapter 7 Outline I. Nervous system function II. Central and peripheral nervous system III. Nervous system cells IV. Myelinated neurons V. Nerve signal transmission VI. Nerve Synapse Copyright © 2009 Pearson Education, Inc. Nervous Tissues . Nervous tissue functions to conduct messages throughout the body. When nerve cells are stimulated, an electrical signal quickly travels through the nerve cell to the nerve ending, triggering events. Copyright © 2009 Pearson Education, Inc. Nervous System . Includes nervous tissue and sensory organs. Nervous system functions to: . Sense the environment – it receives information from both outside and inside the body. Process the information it receives. Respond to information – send out orders. Copyright © 2009 Pearson Education, Inc. Two Parts of the Nervous System 1. Central Nervous System (CNS) . Brain and Spinal Cord. 2. Peripheral Nervous System (PNS) . Nervous tissue outside brain and spine. Sense organs. Copyright © 2009 Pearson Education, Inc. Central Nervous System Peripheral Copyright © 2009 Pearson Education, Inc. Figure 8.1 The nervous system Copyright © 2009 Pearson Education, Inc. Nervous System Cells . Two types of nervous tissue cells. Neurons – The cells that are responsible for transmitting messages. Neuroglial Cells – Cells that support the neurons. Copyright © 2009 Pearson Education, Inc. Neuroglial Cells . Microglia – Immune system cells, engulf bacteria and cellular debris. Astrocytes – Provide nutrients to neurons. Oligodenrocytes and Schwann Cells – Form myelin sheaths. Copyright © 2009 Pearson Education, Inc. Copyright © 2009 Pearson Education, Inc. Parts of a Neuron . Cell body – contains the nucleus, main body of cell. Dendrites – projections from the cell body that carry messages to the cell body. Axon – one projection that carries messages away from the cell body (can be very long). -
Transcriptional and Imaging-Genetic Association of Cortical Interneurons, Brain Function, and Schizophrenia Risk
ARTICLE https://doi.org/10.1038/s41467-020-16710-x OPEN Transcriptional and imaging-genetic association of cortical interneurons, brain function, and schizophrenia risk Kevin M. Anderson 1, Meghan A. Collins1, Rowena Chin1, Tian Ge2,3, Monica D. Rosenberg 1,4 & ✉ Avram J. Holmes 1,3,5 fl 1234567890():,; Inhibitory interneurons orchestrate information ow across the cortex and are implicated in psychiatric illness. Although interneuron classes have unique functional properties and spatial distributions, the influence of interneuron subtypes on brain function, cortical specialization, and illness risk remains elusive. Here, we demonstrate stereotyped negative correlation of somatostatin and parvalbumin transcripts within human and non-human primates. Cortical distributions of somatostatin and parvalbumin cell gene markers are strongly coupled to regional differences in functional MRI variability. In the general population (n = 9,713), parvalbumin-linked genes account for an enriched proportion of heritable variance in in-vivo functional MRI signal amplitude. Single-marker and polygenic cell deconvolution establish that this relationship is spatially dependent, following the topography of parvalbumin expression in post-mortem brain tissue. Finally, schizophrenia genetic risk is enriched among interneuron-linked genes and predicts cortical signal amplitude in parvalbumin-biased regions. These data indicate that the molecular-genetic basis of brain function is shaped by interneuron-related transcripts and may capture individual differences in schizophrenia risk. 1 Department of Psychology, Yale University, New Haven, CT 06520, USA. 2 Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA 02114, USA. 3 Department of Psychiatry, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114, USA. 4 Department of Psychology, University of Chicago, Chicago, IL 60637, USA.