Introduction to Gaussview and Gaussian
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![Automated Construction of Quantum–Classical Hybrid Models Arxiv:2102.09355V1 [Physics.Chem-Ph] 18 Feb 2021](https://docslib.b-cdn.net/cover/7378/automated-construction-of-quantum-classical-hybrid-models-arxiv-2102-09355v1-physics-chem-ph-18-feb-2021-177378.webp)
Automated Construction of Quantum–Classical Hybrid Models Arxiv:2102.09355V1 [Physics.Chem-Ph] 18 Feb 2021
Automated construction of quantum{classical hybrid models Christoph Brunken and Markus Reiher∗ ETH Z¨urich, Laboratorium f¨urPhysikalische Chemie, Vladimir-Prelog-Weg 2, 8093 Z¨urich, Switzerland February 18, 2021 Abstract We present a protocol for the fully automated construction of quantum mechanical-(QM){ classical hybrid models by extending our previously reported approach on self-parametri- zing system-focused atomistic models (SFAM) [J. Chem. Theory Comput. 2020, 16 (3), 1646{1665]. In this QM/SFAM approach, the size and composition of the QM region is evaluated in an automated manner based on first principles so that the hybrid model describes the atomic forces in the center of the QM region accurately. This entails the au- tomated construction and evaluation of differently sized QM regions with a bearable com- putational overhead that needs to be paid for automated validation procedures. Applying SFAM for the classical part of the model eliminates any dependence on pre-existing pa- rameters due to its system-focused quantum mechanically derived parametrization. Hence, QM/SFAM is capable of delivering a high fidelity and complete automation. Furthermore, since SFAM parameters are generated for the whole system, our ansatz allows for a con- venient re-definition of the QM region during a molecular exploration. For this purpose, a local re-parametrization scheme is introduced, which efficiently generates additional clas- sical parameters on the fly when new covalent bonds are formed (or broken) and moved to the classical region. arXiv:2102.09355v1 [physics.chem-ph] 18 Feb 2021 ∗Corresponding author; e-mail: [email protected] 1 1 Introduction In contrast to most protocols of computational quantum chemistry that consider isolated molecules, chemical processes can take place in a vast variety of complex environments. -

Starting SCF Calculations by Superposition of Atomic Densities
Starting SCF Calculations by Superposition of Atomic Densities J. H. VAN LENTHE,1 R. ZWAANS,1 H. J. J. VAN DAM,2 M. F. GUEST2 1Theoretical Chemistry Group (Associated with the Department of Organic Chemistry and Catalysis), Debye Institute, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands 2CCLRC Daresbury Laboratory, Daresbury WA4 4AD, United Kingdom Received 5 July 2005; Accepted 20 December 2005 DOI 10.1002/jcc.20393 Published online in Wiley InterScience (www.interscience.wiley.com). Abstract: We describe the procedure to start an SCF calculation of the general type from a sum of atomic electron densities, as implemented in GAMESS-UK. Although the procedure is well known for closed-shell calculations and was already suggested when the Direct SCF procedure was proposed, the general procedure is less obvious. For instance, there is no need to converge the corresponding closed-shell Hartree–Fock calculation when dealing with an open-shell species. We describe the various choices and illustrate them with test calculations, showing that the procedure is easier, and on average better, than starting from a converged minimal basis calculation and much better than using a bare nucleus Hamiltonian. © 2006 Wiley Periodicals, Inc. J Comput Chem 27: 926–932, 2006 Key words: SCF calculations; atomic densities Introduction hrstuhl fur Theoretische Chemie, University of Kahrlsruhe, Tur- bomole; http://www.chem-bio.uni-karlsruhe.de/TheoChem/turbo- Any quantum chemical calculation requires properly defined one- mole/),12 GAMESS(US) (Gordon Research Group, GAMESS, electron orbitals. These orbitals are in general determined through http://www.msg.ameslab.gov/GAMESS/GAMESS.html, 2005),13 an iterative Hartree–Fock (HF) or Density Functional (DFT) pro- Spartan (Wavefunction Inc., SPARTAN: http://www.wavefun. -
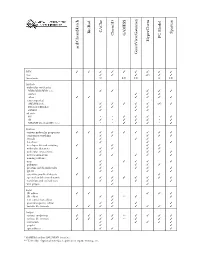
D:\Doc\Workshops\2005 Molecular Modeling\Notebook Pages\Software Comparison\Summary.Wpd
CAChe BioRad Spartan GAMESS Chem3D PC Model HyperChem acd/ChemSketch GaussView/Gaussian WIN TTTT T T T T T mac T T T (T) T T linux/unix U LU LU L LU Methods molecular mechanics MM2/MM3/MM+/etc. T T T T T Amber T T T other TT T T T T semi-empirical AM1/PM3/etc. T T T T T (T) T Extended Hückel T T T T ZINDO T T T ab initio HF * * T T T * T dft T * T T T * T MP2/MP4/G1/G2/CBS-?/etc. * * T T T * T Features various molecular properties T T T T T T T T T conformer searching T T T T T crystals T T T data base T T T developer kit and scripting T T T T molecular dynamics T T T T molecular interactions T T T T movies/animations T T T T T naming software T nmr T T T T T polymers T T T T proteins and biomolecules T T T T T QSAR T T T T scientific graphical objects T T spectral and thermodynamic T T T T T T T T transition and excited state T T T T T web plugin T T Input 2D editor T T T T T 3D editor T T ** T T text conversion editor T protein/sequence editor T T T T various file formats T T T T T T T T Output various renderings T T T T ** T T T T various file formats T T T T ** T T T animation T T T T T graphs T T spreadsheet T T T * GAMESS and/or GAUSSIAN interface ** Text only. -
![Modern Quantum Chemistry with [Open]Molcas](https://docslib.b-cdn.net/cover/4742/modern-quantum-chemistry-with-open-molcas-744742.webp)
Modern Quantum Chemistry with [Open]Molcas
Modern quantum chemistry with [Open]Molcas Cite as: J. Chem. Phys. 152, 214117 (2020); https://doi.org/10.1063/5.0004835 Submitted: 17 February 2020 . Accepted: 11 May 2020 . Published Online: 05 June 2020 Francesco Aquilante , Jochen Autschbach , Alberto Baiardi , Stefano Battaglia , Veniamin A. Borin , Liviu F. Chibotaru , Irene Conti , Luca De Vico , Mickaël Delcey , Ignacio Fdez. Galván , Nicolas Ferré , Leon Freitag , Marco Garavelli , Xuejun Gong , Stefan Knecht , Ernst D. Larsson , Roland Lindh , Marcus Lundberg , Per Åke Malmqvist , Artur Nenov , Jesper Norell , Michael Odelius , Massimo Olivucci , Thomas B. Pedersen , Laura Pedraza-González , Quan M. Phung , Kristine Pierloot , Markus Reiher , Igor Schapiro , Javier Segarra-Martí , Francesco Segatta , Luis Seijo , Saumik Sen , Dumitru-Claudiu Sergentu , Christopher J. Stein , Liviu Ungur , Morgane Vacher , Alessio Valentini , and Valera Veryazov J. Chem. Phys. 152, 214117 (2020); https://doi.org/10.1063/5.0004835 152, 214117 © 2020 Author(s). The Journal ARTICLE of Chemical Physics scitation.org/journal/jcp Modern quantum chemistry with [Open]Molcas Cite as: J. Chem. Phys. 152, 214117 (2020); doi: 10.1063/5.0004835 Submitted: 17 February 2020 • Accepted: 11 May 2020 • Published Online: 5 June 2020 Francesco Aquilante,1,a) Jochen Autschbach,2,b) Alberto Baiardi,3,c) Stefano Battaglia,4,d) Veniamin A. Borin,5,e) Liviu F. Chibotaru,6,f) Irene Conti,7,g) Luca De Vico,8,h) Mickaël Delcey,9,i) Ignacio Fdez. Galván,4,j) Nicolas Ferré,10,k) Leon Freitag,3,l) Marco Garavelli,7,m) Xuejun Gong,11,n) Stefan Knecht,3,o) Ernst D. Larsson,12,p) Roland Lindh,4,q) Marcus Lundberg,9,r) Per Åke Malmqvist,12,s) Artur Nenov,7,t) Jesper Norell,13,u) Michael Odelius,13,v) Massimo Olivucci,8,14,w) Thomas B. -

Benchmarking and Application of Density Functional Methods In
BENCHMARKING AND APPLICATION OF DENSITY FUNCTIONAL METHODS IN COMPUTATIONAL CHEMISTRY by BRIAN N. PAPAS (Under Direction the of Henry F. Schaefer III) ABSTRACT Density Functional methods were applied to systems of chemical interest. First, the effects of integration grid quadrature choice upon energy precision were documented. This was done through application of DFT theory as implemented in five standard computational chemistry programs to a subset of the G2/97 test set of molecules. Subsequently, the neutral hydrogen-loss radicals of naphthalene, anthracene, tetracene, and pentacene and their anions where characterized using five standard DFT treatments. The global and local electron affinities were computed for the twelve radicals. The results for the 1- naphthalenyl and 2-naphthalenyl radicals were compared to experiment, and it was found that B3LYP appears to be the most reliable functional for this type of system. For the larger systems the predicted site specific adiabatic electron affinities of the radicals are 1.51 eV (1-anthracenyl), 1.46 eV (2-anthracenyl), 1.68 eV (9-anthracenyl); 1.61 eV (1-tetracenyl), 1.56 eV (2-tetracenyl), 1.82 eV (12-tetracenyl); 1.93 eV (14-pentacenyl), 2.01 eV (13-pentacenyl), 1.68 eV (1-pentacenyl), and 1.63 eV (2-pentacenyl). The global minimum for each radical does not have the same hydrogen removed as the global minimum for the analogous anion. With this in mind, the global (or most preferred site) adiabatic electron affinities are 1.37 eV (naphthalenyl), 1.64 eV (anthracenyl), 1.81 eV (tetracenyl), and 1.97 eV (pentacenyl). In later work, ten (scandium through zinc) homonuclear transition metal trimers were studied using one DFT 2 functional. -

A Summary of ERCAP Survey of the Users of Top Chemistry Codes
A survey of codes and algorithms used in NERSC chemical science allocations Lin-Wang Wang NERSC System Architecture Team Lawrence Berkeley National Laboratory We have analyzed the codes and their usages in the NERSC allocations in chemical science category. This is done mainly based on the ERCAP NERSC allocation data. While the MPP hours are based on actually ERCAP award for each account, the time spent on each code within an account is estimated based on the user’s estimated partition (if the user provided such estimation), or based on an equal partition among the codes within an account (if the user did not provide the partition estimation). Everything is based on 2007 allocation, before the computer time of Franklin machine is allocated. Besides the ERCAP data analysis, we have also conducted a direct user survey via email for a few most heavily used codes. We have received responses from 10 users. The user survey not only provide us with the code usage for MPP hours, more importantly, it provides us with information on how the users use their codes, e.g., on which machine, on how many processors, and how long are their simulations? We have the following observations based on our analysis. (1) There are 48 accounts under chemistry category. This is only second to the material science category. The total MPP allocation for these 48 accounts is 7.7 million hours. This is about 12% of the 66.7 MPP hours annually available for the whole NERSC facility (not accounting Franklin). The allocation is very tight. The majority of the accounts are only awarded less than half of what they requested for. -

The Molpro Quantum Chemistry Package
The Molpro Quantum Chemistry package Hans-Joachim Werner,1, a) Peter J. Knowles,2, b) Frederick R. Manby,3, c) Joshua A. Black,1, d) Klaus Doll,1, e) Andreas Heßelmann,1, f) Daniel Kats,4, g) Andreas K¨ohn,1, h) Tatiana Korona,5, i) David A. Kreplin,1, j) Qianli Ma,1, k) Thomas F. Miller, III,6, l) Alexander Mitrushchenkov,7, m) Kirk A. Peterson,8, n) Iakov Polyak,2, o) 1, p) 2, q) Guntram Rauhut, and Marat Sibaev 1)Institut f¨ur Theoretische Chemie, Universit¨at Stuttgart, Pfaffenwaldring 55, 70569 Stuttgart, Germany 2)School of Chemistry, Cardiff University, Main Building, Park Place, Cardiff CF10 3AT, United Kingdom 3)School of Chemistry, University of Bristol, Cantock’s Close, Bristol BS8 1TS, United Kingdom 4)Max-Planck Institute for Solid State Research, Heisenbergstraße 1, 70569 Stuttgart, Germany 5)Faculty of Chemistry, University of Warsaw, L. Pasteura 1 St., 02-093 Warsaw, Poland 6)Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California 91125, United States 7)MSME, Univ Gustave Eiffel, UPEC, CNRS, F-77454, Marne-la- Vall´ee, France 8)Washington State University, Department of Chemistry, Pullman, WA 99164-4630 1 Molpro is a general purpose quantum chemistry software package with a long devel- opment history. It was originally focused on accurate wavefunction calculations for small molecules, but now has many additional distinctive capabilities that include, inter alia, local correlation approximations combined with explicit correlation, highly efficient implementations of single-reference correlation methods, robust and efficient multireference methods for large molecules, projection embedding and anharmonic vibrational spectra. -

Computational Chemistry
Computational Chemistry as a Teaching Aid Melissa Boule Aaron Harshman Rexford Hoadley An Interactive Qualifying Project Report Submitted to the Faculty of the Worcester Polytechnic Institute In partial fulfillment of the requirements for the Bachelor of Science Degree Approved by: Professor N. Aaron Deskins 1i Abstract Molecular modeling software has transformed the capabilities of researchers. Molecular modeling software has the potential to be a helpful teaching tool, though as of yet its efficacy as a teaching technique has yet to be proven. This project focused on determining the effectiveness of a particular molecular modeling software in high school classrooms. Our team researched what topics students struggled with, surveyed current high school chemistry teachers, chose a modeling software, and then developed a lesson plan around these topics. Lastly, we implemented the lesson in two separate high school classrooms. We concluded that these programs show promise for the future, but with the current limitations of high school technology, these tools may not be as impactful. 2ii Acknowledgements This project would not have been possible without the help of many great educators. Our advisor Professor N. Aaron Deskins provided driving force to ensure the research ran smoothly and efficiently. Mrs. Laurie Hanlan and Professor Drew Brodeur provided insight and guidance early in the development of our topic. Mr. Mark Taylor set-up our WebMO server and all our other technology needs. Lastly, Mrs. Pamela Graves and Mr. Eric Van Inwegen were instrumental by allowing us time in their classrooms to implement our lesson plans. Without the support of this diverse group of people this project would never have been possible. -

Quantum Chemical Calculations of NMR Parameters
Quantum Chemical Calculations of NMR Parameters Tatyana Polenova University of Delaware Newark, DE Winter School on Biomolecular NMR January 20-25, 2008 Stowe, Vermont OUTLINE INTRODUCTION Relating NMR parameters to geometric and electronic structure Classical calculations of EFG tensors Molecular properties from quantum chemical calculations Quantum chemistry methods DENSITY FUNCTIONAL THEORY FOR CALCULATIONS OF NMR PARAMETERS Introduction to DFT Software Practical examples Tutorial RELATING NMR OBSERVABLES TO MOLECULAR STRUCTURE NMR Spectrum NMR Parameters Local geometry Chemical structure (reactivity) I. Calculation of experimental NMR parameters Find unique solution to CQ, Q, , , , , II. Theoretical prediction of fine structure constants from molecular geometry Classical electrostatic model (EFG)- only in simple ionic compounds Quantum mechanical calculations (Density Functional Theory) (EFG, CSA) ELECTRIC FIELD GRADIENT (EFG) TENSOR: POINT CHARGE MODEL EFG TENSOR IS DETERMINED BY THE COMBINED ELECTRONIC AND NUCLEAR WAVEFUNCTION, NO ANALYTICAL EXPRESSION IN THE GENERAL CASE THE SIMPLEST APPROXIMATION: CLASSICAL POINT CHARGE MODEL n Zie 4 V2,k = 3 Y2,k ()i,i i=1 di 5 ATOMS CONTRIBUTING TO THE EFG TENSOR ARE TREATED AS POINT CHARGES, THE RESULTING EFG TENSOR IS THE SUM WITH RESPECT TO ALL ATOMS VERY CRUDE MODEL, WORKS QUANTITATIVELY ONLY IN SIMPLEST IONIC SYSTEMS, BUT YIELDS QUALITATIVE TRENDS AND GENERAL UNDERSTANDING OF THE SYMMETRY AND MAGNITUDE OF THE EXPECTED TENSOR ELECTRIC FIELD GRADIENT (EFG) TENSOR: POINT CHARGE MODEL n Zie 4 V2,k = 3 Y2,k ()i,i i=1 di 5 Ze V = ; V = 0; V = 0 2,0 d 3 2,±1 2,±2 2Ze V = ; V = 0; V = 0 2,0 d 3 2,±1 2,±2 3 Ze V = ; V = 0; V = 0 2,0 2 d 3 2,±1 2,±2 V2,0 = 0; V2,±1 = 0; V2,±2 = 0 MOLECULAR PROPERTIES FROM QUANTUM CHEMICAL CALCULATIONS H = E See for example M. -

Chem3d 17.0 User Guide Chem3d 17.0
Chem3D 17.0 User Guide Chem3D 17.0 Table of Contents Recent Additions viii Chapter 1: About Chem3D 1 Additional computational engines 1 Serial numbers and technical support 3 About Chem3D Tutorials 3 Chapter 2: Chem3D Basics 5 Getting around 5 User interface preferences 9 Background settings 10 Sample files 10 Saving to Dropbox 10 Chapter 3: Basic Model Building 12 Default settings 12 Selecting a display mode 12 Using bond tools 13 Using the ChemDraw panel 15 Using other 2D drawing packages 15 Building from text 16 Adding fragments 18 Selecting atoms and bonds 18 Atom charges 21 Object position 23 Substructures 24 Refining models 27 Copying and printing 29 Finding structures online 32 Chapter 4: Displaying Models 35 © Copyright 1998-2017 PerkinElmer Informatics Inc., All rights reserved. ii Chem3D 17.0 Display modes 35 Atom and bond size 37 Displaying dot surfaces 38 Serial numbers 38 Displaying atoms 39 Atom symbols 40 Rotating models 41 Atom and bond properties 44 Showing hydrogen bonds 45 Hydrogens and lone pairs 46 Translating models 47 Scaling models 47 Aligning models 47 Applying color 49 Model Explorer 52 Measuring molecules 59 Comparing models by overlay 62 Molecular surfaces 63 Using stereo pairs 72 Stereo enhancement 72 Setting view focus 73 Chapter 5: Building Advanced Models 74 Dummy bonds and dummy atoms 74 Substructures 75 Bonding by proximity 78 Setting measurements 78 Atom and building types 81 Stereochemistry 85 © Copyright 1998-2017 PerkinElmer Informatics Inc., All rights reserved. iii Chem3D 17.0 Building with Cartesian -

FIESTA: the Filipino Initiative on Electronic Structure Theory and Applications
FIESTA: the Filipino Initiative on Electronic Structure Theory and Applications Han Ung Lee, Hayan Lee and Wilfredo Credo Chung* Department of Chemistry, De La Salle University – Manila, 2401 Taft Avenue, Manila, 1004 Philippines *E-mail: [email protected] FIESTA—the Filipino Initiative on Electronic Structure Theory and Applications—a new computational chemistry software is developed. The new software is capable of doing ground- state self-consistent field (SCF) single-point restricted Hartree-Fock (RHF) calculation of polyelectronic and polyatomic systems using the Slater-type orbital basis set STO-3G. The new program implements well-known quantum mechanical theories for practical calculations. FIESTA is written using two programming languages, namely C and FORTRAN. It is accurate and user-friendly. It runs efficiently under the Linux operating system and is able to reproduce the energies calculated using well-established standard quantum chemical software products Gaussian, Firefly and Molpro. To our knowledge, FIESTA is the first and only molecular modeling software developed in the Philippines to date. The software will be extended to calculate the properties of systems such as atoms, molecules, ions and formula units using more sophisticated basis sets and quantum mechanical techniques. Computational chemistry is used for precise gas-phase system properties such as the electronic chemical predictions and refined simulations of energy, orbital energies, nuclear repulsion energy chemical processes with the use of computer and the total energy. programs. Computational data may not yield the At present, there is a good deal of computational exact experimental value from measurements, but it chemistry softwares available. Most of them can predict chemical phenomena qualitatively, and require a commercial license. -

Scale QM/MM-MD Simulations and Its Application to Biomolecular Systems
PUPIL: a Software Integration System for Multi- scale QM/MM-MD Simulations and its Application to Biomolecular Systems Juan Torras, 1 * B.P. Roberts, 2 G.M. Seabra, 3and S.B. Trickey 4 * 1Department of Chemical Engineering, EEI, Universitat Politècnica de Catalunya, Av. Pla de la Massa 8, Igualada 08700, Spain 2 Centre for eResearch, The University of Auckland, Private Bag 92019, Auckland 1142 New Zealand 3Departamento de Química Fundamental, Universidade Federal de Pernambuco, Cidade Universitária, Recife, Pernambuco CEP 50.740-540, Brazil 4Department of Physics, Department of Chemistry, and Quantum Theory Project, Univ. of Florida, Gainesville Florida 32611-8435, USA * Corresponding Authors: [email protected] and [email protected] 1 Abstract PUPIL (Program for User Package Interfacing and Linking) implements a distinctive multi-scale approach to hybrid quantum mechanical/molecular mechanical molecular dynamics (QM/MM-MD) simulations. Originally developed to interface different external programs for multi-scale simulation with applications in the materials sciences, PUPIL is finding increasing use in the study of complex biological systems. Advanced MD techniques from the external packages can be applied readily to a hybrid QM/MM treatment in which the forces and energy for the QM region can be computed by any of the QM methods available in any of the other external packages. Here we give a survey of PUPIL design philosophy, main features, and key implementation decisions, with an orientation to biomolecular simulation. We discuss recently implemented features which enable highly realistic simulations of complex biological systems which have more than one active site that must be treated concurrently.