© Ferrata Storti Foundation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Epigenetic Mechanisms Are Involved in the Oncogenic Properties of ZNF518B in Colorectal Cancer
Epigenetic mechanisms are involved in the oncogenic properties of ZNF518B in colorectal cancer Francisco Gimeno-Valiente, Ángela L. Riffo-Campos, Luis Torres, Noelia Tarazona, Valentina Gambardella, Andrés Cervantes, Gerardo López-Rodas, Luis Franco and Josefa Castillo SUPPLEMENTARY METHODS 1. Selection of genomic sequences for ChIP analysis To select the sequences for ChIP analysis in the five putative target genes, namely, PADI3, ZDHHC2, RGS4, EFNA5 and KAT2B, the genomic region corresponding to the gene was downloaded from Ensembl. Then, zoom was applied to see in detail the promoter, enhancers and regulatory sequences. The details for HCT116 cells were then recovered and the target sequences for factor binding examined. Obviously, there are not data for ZNF518B, but special attention was paid to the target sequences of other zinc-finger containing factors. Finally, the regions that may putatively bind ZNF518B were selected and primers defining amplicons spanning such sequences were searched out. Supplementary Figure S3 gives the location of the amplicons used in each gene. 2. Obtaining the raw data and generating the BAM files for in silico analysis of the effects of EHMT2 and EZH2 silencing The data of siEZH2 (SRR6384524), siG9a (SRR6384526) and siNon-target (SRR6384521) in HCT116 cell line, were downloaded from SRA (Bioproject PRJNA422822, https://www.ncbi. nlm.nih.gov/bioproject/), using SRA-tolkit (https://ncbi.github.io/sra-tools/). All data correspond to RNAseq single end. doBasics = TRUE doAll = FALSE $ fastq-dump -I --split-files SRR6384524 Data quality was checked using the software fastqc (https://www.bioinformatics.babraham. ac.uk /projects/fastqc/). The first low quality removing nucleotides were removed using FASTX- Toolkit (http://hannonlab.cshl.edu/fastxtoolkit/). -

Supplementary Tables S1-S3
Supplementary Table S1: Real time RT-PCR primers COX-2 Forward 5’- CCACTTCAAGGGAGTCTGGA -3’ Reverse 5’- AAGGGCCCTGGTGTAGTAGG -3’ Wnt5a Forward 5’- TGAATAACCCTGTTCAGATGTCA -3’ Reverse 5’- TGTACTGCATGTGGTCCTGA -3’ Spp1 Forward 5'- GACCCATCTCAGAAGCAGAA -3' Reverse 5'- TTCGTCAGATTCATCCGAGT -3' CUGBP2 Forward 5’- ATGCAACAGCTCAACACTGC -3’ Reverse 5’- CAGCGTTGCCAGATTCTGTA -3’ Supplementary Table S2: Genes synergistically regulated by oncogenic Ras and TGF-β AU-rich probe_id Gene Name Gene Symbol element Fold change RasV12 + TGF-β RasV12 TGF-β 1368519_at serine (or cysteine) peptidase inhibitor, clade E, member 1 Serpine1 ARE 42.22 5.53 75.28 1373000_at sushi-repeat-containing protein, X-linked 2 (predicted) Srpx2 19.24 25.59 73.63 1383486_at Transcribed locus --- ARE 5.93 27.94 52.85 1367581_a_at secreted phosphoprotein 1 Spp1 2.46 19.28 49.76 1368359_a_at VGF nerve growth factor inducible Vgf 3.11 4.61 48.10 1392618_at Transcribed locus --- ARE 3.48 24.30 45.76 1398302_at prolactin-like protein F Prlpf ARE 1.39 3.29 45.23 1392264_s_at serine (or cysteine) peptidase inhibitor, clade E, member 1 Serpine1 ARE 24.92 3.67 40.09 1391022_at laminin, beta 3 Lamb3 2.13 3.31 38.15 1384605_at Transcribed locus --- 2.94 14.57 37.91 1367973_at chemokine (C-C motif) ligand 2 Ccl2 ARE 5.47 17.28 37.90 1369249_at progressive ankylosis homolog (mouse) Ank ARE 3.12 8.33 33.58 1398479_at ryanodine receptor 3 Ryr3 ARE 1.42 9.28 29.65 1371194_at tumor necrosis factor alpha induced protein 6 Tnfaip6 ARE 2.95 7.90 29.24 1386344_at Progressive ankylosis homolog (mouse) -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -
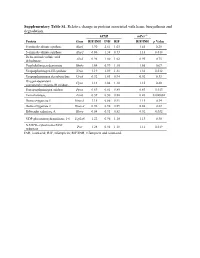
Supplementary Table S1. Relative Change in Proteins Associated with Heme Biosynthesis and Degradation
Supplementary Table S1. Relative change in proteins associated with heme biosynthesis and degradation. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value 5-aminolevulinate synthase Alas1 1.90 2.61 1.05 1.41 0.28 5-aminolevulinate synthase Alas2 0.86 1.38 0.73 1.18 0.018 Delta-aminolevulinic acid Alad 0.96 1.00 1.02 0.95 0.75 dehydratase Porphobilinogen deaminase Hmbs 1.04 0.99 1.10 1.05 0.67 Uroporphyrinogen-III synthase Uros 1.19 1.09 1.31 1.38 0.012 Uroporphyrinogen decarboxylase Urod 0.92 1.03 0.94 0.92 0.33 Oxygen-dependent Cpox 1.13 1.04 1.18 1.15 0.20 coproporphyrinogen-III oxidase, Protoporphyrinogen oxidase Ppox 0.69 0.81 0.85 0.83 0.013 Ferrochelatase, Fech 0.39 0.50 0.88 0.43 0.000002 Heme oxygenase 1 Hmox1 1.15 0.86 0.91 1.11 0.34 Heme oxygenase 2 Hmox2 0.96 0.98 0.89 0.88 0.22 Biliverdin reductase A Blvra 0.84 0.92 0.82 0.92 0.032 UDP-glucuronosyltransferase 1-6 Ugt1a6 1.22 0.96 1.10 1.13 0.30 NADPH--cytochrome P450 Por 1.28 0.92 1.18 1.12 0.019 reductase INH, isoniazid; RIF, rifampicin; RIF/INH, rifampicin and isoniazid. Supplementary Table S2. Relative change in protein nuclear receptors. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value Aryl hydrocarbon receptor Ahr 1.09 0.91 1.00 1.26 0.092 Hepatocyte nuclear factor Hnf1a 0.87 0.97 0.82 0.79 0.027 1-alpha Hepatocyte nuclear factor Hnf4a 0.95 1.05 0.97 1.08 0.20 4-alpha Oxysterols receptor LXR- Nr1h3 0.94 1.16 1.03 1.02 0.42 alpha Bile acid receptor Nr1h4 1.05 1.17 0.98 1.19 0.12 Retinoic acid receptor Rxra 0.88 1.03 0.83 0.95 0.12 RXR-alpha Peroxisome proliferator- -
P4 Atpases: Flippases in Health and Disease
Int. J. Mol. Sci. 2013, 14, 7897-7922; doi:10.3390/ijms14047897 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Review P4 ATPases: Flippases in Health and Disease Vincent A. van der Mark *, Ronald P.J. Oude Elferink and Coen C. Paulusma Tytgat Institute for Liver and Intestinal Research, Academic Medical Center, Meibergdreef 69-71, 1105 BK Amsterdam, The Netherlands; E-Mails: [email protected] (R.P.J.O.E.); [email protected] (C.C.P.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +31-205-668-156; Fax: +31-205-669-190. Received: 6 March 2013; in revised form: 28 March 2013 / Accepted: 7 April 2013 / Published: 11 April 2013 Abstract: P4 ATPases catalyze the translocation of phospholipids from the exoplasmic to the cytosolic leaflet of biological membranes, a process termed “lipid flipping”. Accumulating evidence obtained in lower eukaryotes points to an important role for P4 ATPases in vesicular protein trafficking. The human genome encodes fourteen P4 ATPases (fifteen in mouse) of which the cellular and physiological functions are slowly emerging. Thus far, deficiencies of at least two P4 ATPases, ATP8B1 and ATP8A2, are the cause of severe human disease. However, various mouse models and in vitro studies are contributing to our understanding of the cellular and physiological functions of P4-ATPases. This review summarizes current knowledge on the basic function of these phospholipid translocating proteins, their proposed action in intracellular vesicle transport and their physiological role. -

Phospholipid Flippases in B Cells and Platelets
Phospholipid Flippases in B cells and Platelets Weidong Jing July 2019 A thesis submitted for the degree of Doctor of Philosophy of The Australian National University © Copyright by Weidong Jing 2019 All Rights Reserved Statement The research presented in this thesis is my own original work except where stated otherwise. -------------------------------- Weidong Jing i Acknowledgments First of all, I would like to express my deepest gratitude to my supervisor, Professor Stefan Bröer, who gave me a lot of beneficial guidance, generous help and support in the process of completing my study. He gave the freedom to explore on my own, and also kept me on the track at every step of my PhD. Then I would like to thank Ms Angelika Bröer, the research officer of our lab, for her careful guidance and selfless help in experimental techniques. I also want to thank all the members of the laboratory, no matter they have left or are still here, they have provided me with the help within their power in the experiment and brought me a lot of joy in life. Thanks also go to our divisional STO Dr. Farid Rahimi for his generous helps during my PhD. I also would to thank my co-supervisor Associate Professor Anselm Enders from JCSMR, for his help in mice work and his constructive suggestions and ideas on this flippase project. Many thanks go to Dr. Mehmet Yabas, who helped me a lot on flow cytometry and B cell works. Also, many thanks go to Associate Professor Elizabeth Gardiner and Dr. Lucy Coupland from JCSMR, who are experts on platelets biology, they gave me quite a lot of valuable suggestions and generous help on platelet experiments. -

D-Serine, an Emerging Biomarker of Kidney Diseases, Is a Hidden Substrate of Sodium-Coupled Monocarboxylate Transporters
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.10.244822; this version posted August 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Wiriyasermkul, et al. 1 D-Serine, an emerging biomarker of kidney diseases, is a hidden substrate of 2 sodium-coupled monocarboxylate transporters 3 4 Pattama Wiriyasermkul1, Satomi Moriyama1, Yoko Tanaka1, Pornparn Kongpracha1, Nodoka 5 Nakamae1, Masataka Suzuki2, Tomonori Kimura3,4, Masashi Mita5, Jumpei Sasabe2, Shushi 6 Nagamori1,# 7 8 1Department of Collaborative Research for Biomolecular Dynamics, Nara Medical 9 University, Nara, Japan 10 2Department of Pharmacology, Keio University School of Medicine, Tokyo, Japan 11 3KAGAMI Project, 4Reverse Translational Research Project, Center for Rare Disease 12 Research, National Institutes of Biomedical Innovation, Health and Nutrition (NIBIOHN). 13 Osaka, Japan 14 5KAGAMI Inc., Osaka, Japan 15 16 # Corresponding contact information: [email protected] 17 18 19 20 21 Keywords: transporter, proteome, ischemia-reperfusion injury (IRI), acute kidney injury 22 (AKI), chronic kidney diseases (CKD), membrane 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.10.244822; this version posted August 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Wiriyasermkul, et al. 23 Abstract 24 Delay in diagnosis of renal injury has profound impacts on morbidity and mortality. -

Proteomic Landscape of the Human Choroid–Retinal Pigment Epithelial Complex
Supplementary Online Content Skeie JM, Mahajan VB. Proteomic landscape of the human choroid–retinal pigment epithelial complex. JAMA Ophthalmol. Published online July 24, 2014. doi:10.1001/jamaophthalmol.2014.2065. eFigure 1. Choroid–retinal pigment epithelial (RPE) proteomic analysis pipeline. eFigure 2. Gene ontology (GO) distributions and pathway analysis of human choroid– retinal pigment epithelial (RPE) protein show tissue similarity. eMethods. Tissue collection, mass spectrometry, and analysis. eTable 1. Complete table of proteins identified in the human choroid‐RPE using LC‐ MS/MS. eTable 2. Top 50 signaling pathways in the human choroid‐RPE using MetaCore. eTable 3. Top 50 differentially expressed signaling pathways in the human choroid‐RPE using MetaCore. eTable 4. Differentially expressed proteins in the fovea, macula, and periphery of the human choroid‐RPE. eTable 5. Differentially expressed transcription proteins were identified in foveal, macular, and peripheral choroid‐RPE (p<0.05). eTable 6. Complement proteins identified in the human choroid‐RPE. eTable 7. Proteins associated with age related macular degeneration (AMD). This supplementary material has been provided by the authors to give readers additional information about their work. © 2014 American Medical Association. All rights reserved. 1 Downloaded From: https://jamanetwork.com/ on 09/25/2021 eFigure 1. Choroid–retinal pigment epithelial (RPE) proteomic analysis pipeline. A. The human choroid‐RPE was dissected into fovea, macula, and periphery samples. B. Fractions of proteins were isolated and digested. C. The peptide fragments were analyzed using multi‐dimensional LC‐MS/MS. D. X!Hunter, X!!Tandem, and OMSSA were used for peptide fragment identification. E. Proteins were further analyzed using bioinformatics. -

Data Set 1. Biological Analysis of the Genes Found to Be Significant in the Endotoxin Study
Data Set 1. Biological analysis of the genes found to be significant in the endotoxin study. Pages 2 – 105: Q values, gene names and annotation on the genes significant at FDR = 0.1%. Pages 106 – 127: Global functional analysis of the down-regulated genes. Pages 128 – 169: Global functional analysis of the up-regulated genes. Probe Set q-value Direction Gene Annotation 117_at 9.60E-05 up HSPA6 heat shock 70kDa protein 6 (HSP70B') 1405_i_at 2.00E-06 down CCL5 chemokine (C-C motif) ligand 5 PRP8 pre-mRNA processing factor 8 200000_s_at 2.50E-05 down PRPF8 homolog (yeast) PRP8 pre-mRNA processing factor 8 200000_s_at 0.000712 down PRPF8 homolog (yeast) 200001_at 0.000658 down CAPNS1 calpain, small subunit 1 200002_at 2.00E-06 down RPL35 ribosomal protein L35 200002_at 2.00E-06 down RPL35 ribosomal protein L35 200003_s_at 1.10E-05 down RPL28 ribosomal protein L28 200003_s_at 1.10E-05 down RPL28 ribosomal protein L28 eukaryotic translation initiation factor 3, 200005_at 2.00E-06 down EIF3S7 subunit 7 zeta, 66/67kDa eukaryotic translation initiation factor 3, 200005_at 2.00E-06 down EIF3S7 subunit 7 zeta, 66/67kDa Parkinson disease (autosomal 200006_at 4.70E-05 down PARK7 recessive, early onset) 7 Parkinson disease (autosomal 200006_at 8.70E-05 down PARK7 recessive, early onset) 7 200008_s_at 5.40E-05 up GDI2 GDP dissociation inhibitor 2 200008_s_at 0.000361 up GDI2 GDP dissociation inhibitor 2 200009_at 0.000171 down GDI2 GDP dissociation inhibitor 2 200010_at 2.00E-06 down RPL11 ribosomal protein L11 200010_at 2.00E-06 down RPL11 ribosomal -

Proteomic Analysis and Functional Characterization of P4-Atpase
www.nature.com/scientificreports OPEN Proteomic Analysis and Functional Characterization of P4-ATPase Phospholipid Flippases from Murine Received: 29 March 2018 Accepted: 5 July 2018 Tissues Published: xx xx xxxx Jiao Wang1,2, Laurie L. Molday1, Theresa Hii1, Jonathan A. Coleman1, Tieqiao Wen2, Jens P. Andersen 3 & Robert S. Molday1 P4-ATPases are a subfamily of P-type ATPases that fip phospholipids across membranes to generate lipid asymmetry, a property vital to many cellular processes. Mutations in several P4-ATPases have been linked to severe neurodegenerative and metabolic disorders. Most P4-ATPases associate with one of three accessory subunit isoforms known as CDC50A (TMEM30A), CDC50B (TMEM30B), and CDC50C (TMEM30C). To identify P4-ATPases that associate with CDC50A, in vivo, and determine their tissue distribution, we isolated P4-ATPases-CDC50A complexes from retina, brain, liver, testes, and kidney on a CDC50A immunoafnity column and identifed and quantifed P4-ATPases from their tryptic peptides by mass spectrometry. Of the 12 P4-ATPase that associate with CDC50 subunits, 10 P4- ATPases were detected. Four P4-ATPases (ATP8A1, ATP11A, ATP11B, ATP11C) were present in all fve tissues. ATP10D was found in low amounts in liver, brain, testes, and kidney, and ATP8A2 was present in signifcant amounts in retina, brain, and testes. ATP8B1 was detected only in liver, ATP8B3 and ATP10A only in testes, and ATP8B2 primarily in brain. We also show that ATP11A, ATP11B and ATP11C, like ATP8A1 and ATP8A2, selectively fip phosphatidylserine and phosphatidylethanolamine across membranes. These studies provide new insight into the tissue distribution, relative abundance, subunit interactions and substrate specifcity of P4-ATPase-CDC50A complexes. -

An Interstitial Deletion-Insertion Involving Chromosomes 2P25.3 and Xq27.1, Near SOX3, Causes X-Linked Recessive Hypoparathyroidism
An interstitial deletion-insertion involving chromosomes 2p25.3 and Xq27.1, near SOX3, causes X-linked recessive hypoparathyroidism Michael R. Bowl, … , Michael P. Whyte, Rajesh V. Thakker J Clin Invest. 2005;115(10):2822-2831. https://doi.org/10.1172/JCI24156. Research Article Genetics X-linked recessive hypoparathyroidism, due to parathyroid agenesis, has been mapped to a 906-kb region on Xq27 that contains 3 genes (ATP11C, U7snRNA, and SOX3), and analyses have not revealed mutations. We therefore characterized this region by combined analysis of single nucleotide polymorphisms and sequence-tagged sites. This identified a 23- to 25-kb deletion, which did not contain genes. However, DNA fiber–FISH and pulsed-field gel electrophoresis revealed an approximately 340-kb insertion that replaced the deleted fragment. Use of flow-sorted X chromosome–specific libraries and DNA sequence analyses revealed that the telomeric and centromeric breakpoints on X were, respectively, approximately 67 kb downstream of SOX3 and within a repetitive sequence. Use of a monochromosomal somatic cell hybrid panel and metaphase-FISH mapping demonstrated that the insertion originated from 2p25 and contained a segment of the SNTG2 gene that lacked an open reading frame. However, the deletion- insertion [del(X)(q27.1) inv ins (X;2)(q27.1;p25.3)], which represents a novel abnormality causing hypoparathyroidism, could result in a position effect on SOX3 expression. Indeed, SOX3 expression was demonstrated, by in situ hybridization, in the developing parathyroid tissue of mouse embryos between 10.5 and 15.5 days post coitum. Thus, our results indicate a likely new role for SOX3 in the embryonic development of the parathyroid glands.