Mtdna Variation in Rattus Exulans from the Chatham and Kermadec Islands
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Memory Work on R ¯Ekohu (Chatham Islands) Kingsley Baird
Memory Connection Volume 3 Number 1 © 2019 The Memory Waka Hokopanopano Ka Toi Moriori (Reigniting Moriori Arts): Memory Work on R ¯ekohu (Chatham Islands) Kingsley Baird Hokopanopano Ka Toi Moriori (Reigniting Moriori Arts): Memory Work on R ¯ekohu (Chatham Islands)—Kingsley Baird Hokopanopano Ka Toi Moriori (Reigniting Moriori Arts): Memory Work on R ¯ekohu (Chatham Islands) Kingsley Baird Abstract Since European discovery of Re¯kohu (Chatham Islands) in 1791, the pacifist Moriori population declined rapidly as a result of introduced diseases (to which they had no immunity) and killing and enslavement by M¯aori iwi (tribes) from the New Zealand ‘mainland’ following their invasion in 1835. When (full-blooded) Tame Horomona Rehe—described on his headstone as the ‘last of the Morioris’— died in 1933, the Moriori were widely considered to be an extinct people. In February 2016, Moriori rangata m¯a tua (elders) and rangatehi (youth), artists and designers, archaeologists, a conservator and an arborist gathered at Ko¯ pinga Marae on Re¯kohu to participate in a w¯a nanga organized by the Hokotehi Moriori Trust. Its purpose was to enlist the combined expertise and commitment of the participants to hokopanopano ka toi Moriori (reignite Moriori arts)—principally those associated with r¯a kau momori (‘carving’ on living ko¯ pi trees)—through discussion, information exchange, speculation, toolmaking and finally, tree carving. In addition to providing a brief cultural and historical background, this paper recounts some of the memory work of the w¯a nanga from the perspective of one of the participants whose fascination for Moriori and the resilience of their culture developed from Michael King’s 1989 book, Moriori: A People Rediscovered. -

Biodiversity of the Kermadec Islands and Offshore Waters of the Kermadec Ridge: Report of a Coastal, Marine Mammal and Deep-Sea Survey (TAN1612)
Biodiversity of the Kermadec Islands and offshore waters of the Kermadec Ridge: report of a coastal, marine mammal and deep-sea survey (TAN1612) New Zealand Aquatic Environment and Biodiversity Report No. 179 Clark, M.R.; Trnski, T.; Constantine, R.; Aguirre, J.D.; Barker, J.; Betty, E.; Bowden, D.A.; Connell, A.; Duffy, C.; George, S.; Hannam, S.; Liggins, L..; Middleton, C.; Mills, S.; Pallentin, A.; Riekkola, L.; Sampey, A.; Sewell, M.; Spong, K.; Stewart, A.; Stewart, R.; Struthers, C.; van Oosterom, L. ISSN 1179-6480 (online) ISSN 1176-9440 (print) ISBN 978-1-77665-481-9 (online) ISBN 978-1-77665-482-6 (print) January 2017 Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries websites at: http://www.mpi.govt.nz/news-resources/publications.aspx http://fs.fish.govt.nz go to Document library/Research reports © Crown Copyright - Ministry for Primary Industries TABLE OF CONTENTS EXECUTIVE SUMMARY 1 1. INTRODUCTION 3 1.1 Objectives: 3 1.2 Objective 1: Benthic offshore biodiversity 3 1.3 Objective 2: Marine mammal research 4 1.4 Objective 3: Coastal biodiversity and connectivity 5 2. METHODS 5 2.1 Survey area 5 2.2 Survey design 6 Offshore Biodiversity 6 Marine mammal sampling 8 Coastal survey 8 Station recording 8 2.3 Sampling operations 8 Multibeam mapping 8 Photographic transect survey 9 Fish and Invertebrate sampling 9 Plankton sampling 11 Catch processing 11 Environmental sampling 12 Marine mammal sampling 12 Dive sampling operations 12 Outreach 13 3. -

My Friend, the Volcano (Adapted from the 2004 Submarine Ring of Fire Expedition)
New Zealand American Submarine Ring of Fire 2007 My Friend, The Volcano (adapted from the 2004 Submarine Ring of Fire Expedition) FOCUS MAXIMUM NUMBER OF STUDENTS Ecological impacts of volcanism in the Mariana 30 and Kermadec Islands KEY WORDS GRADE LEVEL Ring of Fire 5-6 (Life Science/Earth Science) Asthenosphere Lithosphere FOCUS QUESTION Magma What are the ecological impacts of volcanic erup- Fault tions on tropical island arcs? Transform boundary Convergent boundary LEARNING OBJECTIVES Divergent boundary Students will be able to describe at least three Subduction beneficial impacts of volcanic activity on marine Tectonic plate ecosystems. BACKGROUND INFORMATION Students will be able to explain the overall tec- The Submarine Ring of Fire is an arc of active vol- tonic processes that cause volcanic activity along canoes that partially encircles the Pacific Ocean the Mariana Arc and Kermadec Arc. Basin, including the Kermadec and Mariana Islands in the western Pacific, the Aleutian Islands MATERIALS between the Pacific and Bering Sea, the Cascade Copies of “Marianas eruption killed Anatahan’s Mountains in western North America, and numer- corals,” one copy per student or student group ous volcanoes on the western coasts of Central (from http://www.cdnn.info/eco/e030920/e030920.html) America and South America. These volcanoes result from the motion of large pieces of the AUDIO/VISUAL MATERIALS Earth’s crust known as tectonic plates. None Tectonic plates are portions of the Earth’s outer TEACHING TIME crust (the lithosphere) about 5 km thick, as Two or three 45-minute class periods, plus time well as the upper 60 - 75 km of the underlying for student research mantle. -

Patterns of Prehistoric Human Mobility in Polynesia Indicated by Mtdna from the Pacific Rat (Rattus Exulans͞population Mobility)
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 15145–15150, December 1998 Anthropology Patterns of prehistoric human mobility in Polynesia indicated by mtDNA from the Pacific rat (Rattus exulansypopulation mobility) E. MATISOO-SMITH*†,R.M.ROBERTS‡,G.J.IRWIN*, J. S. ALLEN*, D. PENNY§, AND D. M. LAMBERT¶ *Department of Anthropology and ‡School of Biological Sciences, University of Auckland, P. B. 92019 Auckland, New Zealand; and §Molecular Genetics Unit and ¶Department of Ecology, Massey University, P. B. 11222 Palmerston North, New Zealand Communicated by R. C. Green, University of Auckland, Auckland, New Zealand, October 14, 1998 (received for review July 20, 1998) ABSTRACT Human settlement of Polynesia was a major Recent genetic research focusing on Polynesian populations event in world prehistory. Despite the vastness of the distances has contributed significantly to our understanding of the covered, research suggests that prehistoric Polynesian popu- ultimate origins of this last major human migration. Studies of lations maintained spheres of continuing interaction for at globin gene variation (2) and mtDNA lineages of modern least some period of time in some regions. A low level of genetic Polynesians (3, 4) and studies of ancient DNA from Lapita- variation in ancestral Polynesian populations, genetic admix- associated skeletons (5) may indicate that some degree of ture (both prehistoric and post-European contact), and severe admixture with populations in Near Oceania occurred as more population crashes resulting from introduction of European remote biological ancestors left Southeast Asia and passed diseases make it difficult to trace prehistoric human mobility through Near Oceania. An alternative hypothesis is that the in the region by using only human genetic and morphological biological ancestors of these groups were one of a number of markers. -

ARTHROPODA Subphylum Hexapoda Protura, Springtails, Diplura, and Insects
NINE Phylum ARTHROPODA SUBPHYLUM HEXAPODA Protura, springtails, Diplura, and insects ROD P. MACFARLANE, PETER A. MADDISON, IAN G. ANDREW, JOCELYN A. BERRY, PETER M. JOHNS, ROBERT J. B. HOARE, MARIE-CLAUDE LARIVIÈRE, PENELOPE GREENSLADE, ROSA C. HENDERSON, COURTenaY N. SMITHERS, RicarDO L. PALMA, JOHN B. WARD, ROBERT L. C. PILGRIM, DaVID R. TOWNS, IAN McLELLAN, DAVID A. J. TEULON, TERRY R. HITCHINGS, VICTOR F. EASTOP, NICHOLAS A. MARTIN, MURRAY J. FLETCHER, MARLON A. W. STUFKENS, PAMELA J. DALE, Daniel BURCKHARDT, THOMAS R. BUCKLEY, STEVEN A. TREWICK defining feature of the Hexapoda, as the name suggests, is six legs. Also, the body comprises a head, thorax, and abdomen. The number A of abdominal segments varies, however; there are only six in the Collembola (springtails), 9–12 in the Protura, and 10 in the Diplura, whereas in all other hexapods there are strictly 11. Insects are now regarded as comprising only those hexapods with 11 abdominal segments. Whereas crustaceans are the dominant group of arthropods in the sea, hexapods prevail on land, in numbers and biomass. Altogether, the Hexapoda constitutes the most diverse group of animals – the estimated number of described species worldwide is just over 900,000, with the beetles (order Coleoptera) comprising more than a third of these. Today, the Hexapoda is considered to contain four classes – the Insecta, and the Protura, Collembola, and Diplura. The latter three classes were formerly allied with the insect orders Archaeognatha (jumping bristletails) and Thysanura (silverfish) as the insect subclass Apterygota (‘wingless’). The Apterygota is now regarded as an artificial assemblage (Bitsch & Bitsch 2000). -

Obsidian from Macauley Island: a New Zealand Connection
www.aucklandmuseum.com Obsidian from Macauley Island: a New Zealand connection Louise Furey Auckland War Memorial Museum Callan Ross-Sheppard McGill University, Montreal Kath E. Prickett Auckland War Memorial Museum Abstract An obsidian flake collected from Macauley Island during the Kermadec Expedition has been analysed to determine the source. The result indicates a Mayor Island, New Zealand, origin, supporting previous results on obsidian from Raoul Island that Polynesians travelled back into the Pacific from New Zealand. Keywords obsidian; Pacific colonisation; Pacific voyaging; Macauley Island. INTRODUCTION The steep terrain meant there were only a few suitable habitation places confined to flat areas of limited size on The Kermadec Islands comprise Raoul Island, the largest the south and north coasts. The only landing places were of the group, the nearby Meyer Islands and Herald adjacent to these flats. Botanical surveys have recorded Islets, and 100–155 km to the south are Macauley, the presence of Polynesian tropical cultigens including Curtis and Cheeseman iIslands, and L’Esperence Rock. candlenut (Aleurites monuccana) and ti (Cordyline Raoul is approximately 1000 km to the northeast of fruticosa) (Sykes 1977) which are not endemic to the New Zealand. During the 2011 Kermadec Expedition, island and it is assumed they were transported there by samples of naturally occurring obsidian were obtained Polynesian settlers. Polynesian tuberous vegetables such from Raoul, the Meyer Islands, and from the sea floor off as taro (Colocasia esculenta) and kumara (Ipomoea Curtis Island. These are now in the Auckland Museum batatas) were also recorded but their introduction collection. More importantly however was the single attributed to later 19th century immigrants. -

Rekohu Report (2016 Newc).Vp
Rekohu REKOHU AReporton MorioriandNgatiMutungaClaims in the Chatham Islands Wa i 6 4 WaitangiTribunalReport2001 The cover design by Cliff Whiting invokes the signing of the Treaty of Waitangi and the consequent interwoven development of Maori and Pakeha history in New Zealand as it continuously unfoldsinapatternnotyetcompletelyknown AWaitangiTribunalreport isbn 978-1-86956-260-1 © Waitangi Tribunal 2001 Reprinted with corrections 2016 www.waitangi-tribunal.govt.nz Produced by the Waitangi Tribunal Published by Legislation Direct, Wellington, New Zealand Printed by Printlink, Lower Hutt, New Zealand Set in Adobe Minion and Cronos multiple master typefaces e nga mana,e nga reo,e nga karangaranga maha tae noa ki nga Minita o te Karauna. ko tenei te honore,hei tuku atu nga moemoea o ratou i kawea te kaupapa nei. huri noa ki a ratou kua wheturangitia ratou te hunga tautoko i kokiri,i mau ki te kaupapa,mai te timatanga,tae noa ki te puawaitanga o tenei ripoata. ahakoa kaore ano ki a kite ka tangi,ka mihi,ka poroporoakitia ki a ratou. ki era o nga totara o Te-Wao-nui-a-Tane,ki a Te Makarini,ki a Horomona ma ki a koutou kua huri ki tua o te arai haere,haere,haere haere i runga i te aroha,me nga roimata o matou kua mahue nei. e kore koutou e warewaretia. ma te Atua koutou e manaaki,e tiaki ka huri Contents Letter of Transmittal _____________________________________________________xiii 1. Summary 1.1 Background ________________________________________________________1 1.2 Historical Claims ____________________________________________________4 1.3 Contemporary Claims ________________________________________________9 1.4 Preliminary Claims __________________________________________________11 1.5 Rekohu, the Chatham Islands, or Wharekauri? _____________________________12 1.6 Concluding Remarks ________________________________________________13 2. -

Reasons to Visit The
Reasons to visit the First place in the world to greet the new dawn Home of unique Chatham Islands birds and plants Visit significant sites of history and heritage Learn about the ancient Moriori covenant of peace Go fishing and hunting Enjoy rugged and awe inspiring landscapes Meet the people of the Chatham Islands Top 20 “Must See” Attractions Admiral Gardens & Pitt Island Kahukura Studio Point Munning Seal Colony Awatotara Bush Coastal Walking Track Port Hutt Basalt Columns Stone Cottage Chatham Cottage Crafts Sunderland Flying Boat Chatham Island Food Co. Taiko Camp and Gap Sanctuary Chatham Islands Museum Tommy Solomon Eva-Cherie Artz & Memorial Statue Studio 44°s Waitangi West Fishing Charters Wharekauri Station and Kaingaroa Splatter Rock chathamislands.co.nz Kopinga Marae DOC Walks Splatter/Taniwha Rock Skirmish Bay Stay " Wharekauri The Landing Ponga Whare Sunderland Flying Boat Maunganui Stone Cottage Ocean Mail Scenic Reserve Waitangi West " Point Munning " N KAIWHATA RD Conservation Covenant O Kaingaroa PORT R Seal Colony HU T Nikau Bush Port TT RD H R Conservation Area Te Whakaru Graveyard Hutt D " German Missionaries Settlement Basalt Columns Go Wild Nursery Thomas Currell J M Barker (Hapupu) Port Hutt Bay Stays Admiral Garden & Kahukura Studio AIRBASE RD National Historic Reserve Henga Lodge Te Whanga Henga Scenic Reserve Lagoon Chatham Island (R kohu / Wh arekauri) Tikitiki Hill Conservation Area " Chatham Island Charters Te One Pitt Island is Lake Pitt Island Guided Access Only " Huro " Te Matarae Kopinga Marae (Rangihaute/Rangiauria) -
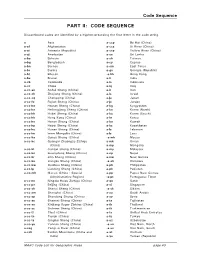
Code Sequence
Code Sequence PART II: CODE SEQUENCE Discontinued codes are identified by a hyphen preceding the first letter in the code string. a Asia a-ccp Bo Hai (China) a-af Afghanistan a-ccs Xi River (China) a-ai Armenia (Republic) a-ccy Yellow River (China) a-aj Azerbaijan a-ce Sri Lanka a-ba Bahrain a-ch Taiwan a-bg Bangladesh a-cy Cyprus a-bn Borneo a-em East Timor a-br Burma a-gs Georgia (Republic) a-bt Bhutan -a-hk Hong Kong a-bx Brunei a-ii India a-cb Cambodia a-io Indonesia a-cc China a-iq Iraq a-cc-an Anhui Sheng (China) a-ir Iran a-cc-ch Zhejiang Sheng (China) a-is Israel a-cc-cq Chongqing (China) a-ja Japan a-cc-fu Fujian Sheng (China) a-jo Jordan a-cc-ha Hainan Sheng (China) a-kg Kyrgyzstan a-cc-he Heilongjiang Sheng (China) a-kn Korea (North) a-cc-hh Hubei Sheng (China) a-ko Korea (South) a-cc-hk Hong Kong (China) a-kr Korea a-cc-ho Henan Sheng (China) a-ku Kuwait a-cc-hp Hebei Sheng (China) a-kz Kazakhstan a-cc-hu Hunan Sheng (China) a-le Lebanon a-cc-im Inner Mongolia (China) a-ls Laos a-cc-ka Gansu Sheng (China) -a-mh Macao a-cc-kc Guangxi Zhuangzu Zizhiqu a-mk Oman (China) a-mp Mongolia a-cc-ki Jiangxi Sheng (China) a-my Malaysia a-cc-kn Guangdong Sheng (China) a-np Nepal a-cc-kr Jilin Sheng (China) a-nw New Guinea a-cc-ku Jiangsu Sheng (China) -a-ok Okinawa a-cc-kw Guizhou Sheng (China) a-ph Philippines a-cc-lp Liaoning Sheng (China) a-pk Pakistan a-cc-mh Macao (China : Special a-pp Papua New Guinea Administrative Region) -a-pt Portuguese Timor a-cc-nn Ningxia Huizu Zizhiqu (China) a-qa Qatar a-cc-pe Beijing (China) a-si Singapore -

Conservation Status of New Zealand Marine Mammals, 2019 Report
2019 NEW ZEALAND THREAT CLASSIFICATION SERIES 29 Conservation status of New Zealand marine mammals, 2019 C.S. Baker, L. Boren, S. Childerhouse, R. Constantine, A. van Helden, D. Lundquist, W. Rayment and J.R. Rolfe Dedication This report is dedicated to the memory of Dr Rob Mattlin, a founding member of this Committee, who passed away in May 2019. Rob’s knowledge, insight and personality will be greatly missed by the Committee and by the marine mammals for which he was so passionate. Rob made New Zealand his home after leaving his native USA to undertake a PhD on New Zealand fur seals at Canterbury University. He proceeded to dedicate his life to marine science with a particular focus on marine mammals and spent most of his time based in New Zealand, apart from a role at the US Marine Mammal Commission. His contribution to our understanding of marine mammal biology and ecology and, in particular, his expertise in interactions with fisheries, was a key factor in achieving many positive outcomes for marine mammals in New Zealand. He made significant contributions to the conservation and management of both New Zealand sea lions and Hector’s dolphins. The Committee would like to recognise the unique balance of wisdom, humour, and tenacity that Rob brought to his work and hope that we can continue to match his enthusiasm and passion. Cover: The southern right whale (Eubalaena australis) population is recovering from perilously low numbers and is now assessed as At Risk – Recovering, an improvement on its previous Threatened status. Photo: Will Rayment. -

Aquatic Vegetation of Chatham Island (Rekohu)
Aquatic vegetation of Chatham Island (Rekohu) DOC SCIENCE INTERNAL SERIES 164 Paul D. Champion and John S. Clayton Published by Department of Conservation PO Box 10-420 Wellington, New Zealand DOC Science Internal Series is a published record of scientific research carried out, or advice given, by Department of Conservation staff or external contractors funded by DOC. It comprises reports and short communications that are peer-reviewed. Individual contributions to the series are first released on the departmental website in pdf form. Hardcopy is printed, bound, and distributed at regular intervals. Titles are also listed in the DOC Science Publishing catalogue on the website, refer http://www.doc.govt.nz under Publications, then Science and Research. © Copyright March 200, New Zealand Department of Conservation ISSN 1175–6519 ISBN 0–478–22086–3 In the interest of forest conservation, DOC Science Publishing supports paperless electronic publishing. When printing, recycled paper is used wherever possible. This is a client report commissioned by Wellington Conservancy and funded from the Science Advice Fund. It was prepared for publication by DOC Science Publishing, Science & Research Unit; editing and layout by Geoff Gregory. Publication was approved by the Manager, Science & Research Unit, Science Technology and Information Services, Department of Conservation, Wellington. CONTENTS Abstract 5 1. Introduction 6 2. Methods 6 3. Results 9 3.1 Characterisation of waterbodies 9 3.2 Aquatic vegetation 9 3.3 Aquatic fauna 11 4. Discussion 11 4.1 Current aquatic weed status 11 4.2 Native aquatic plant species in the lakes 12 4.2.1 New records for the Chatham Islands 13 4.2.2 Species of limited distribution 14 4.3 Fauna in the lakes 14 4.4 Other management issues 15 5. -

Chatham Island
Reasons to visit the First place in the world to greet the new dawn Home of unique Chatham Islands birds and plants Visit significant sites of history and heritage Learn about the ancient Moriori covenant of peace Go fishing and hunting Enjoy rugged and awe inspiring landscapes Meet the people of the Chatham Islands Top 20 “Must See” Attractions Admiral Gardens & Pitt Island Kahukura Studio Point Munning Seal Colony Awatotara Bush Coastal Walking Track Port Hutt Basalt Columns Stone Cottage Chatham Cottage Crafts Sunderland Flying Boat Chatham Island Food Co. Taiko Camp and Gap Sanctuary Chatham Islands Museum Tommy Solomon Eva-Cherie Artz & Memorial Statue Studio 44°s Waitangi West Fishing Charters Wharekauri Station and Kaingaroa Splatter Rock chathamislands.co.nz Kopinga Marae DOC Walks Splatter/Taniwha Rock Skirmish Bay Stay " Wharekauri The Landing Ponga Whare Sunderland Flying Boat Maunganui Stone Cottage Ocean Mail Scenic Reserve Waitangi West " Point Munning " N KAIWHATA RD Conservation Covenant O Kaingaroa PORT R Seal Colony HU T Nikau Bush Port TT RD H R Conservation Area Te Whakaru Graveyard Hutt D " German Missionaries Settlement Basalt Columns Go Wild Nursery Thomas Currell J M Barker (Hapupu) Port Hutt Bay Stays Admiral Garden & Kahukura Studio AIRBASE RD National Historic Reserve Henga Lodge Te Whanga Henga Scenic Reserve Lagoon Chatham Island (R kohu / Wh arekauri) Tikitiki Hill Conservation Area " Chatham Island Charters Te One Pitt Island is Lake Pitt Island Guided Access Only " Huro " Te Matarae Kopinga Marae (Rangihaute/Rangiauria)