Patterns of Prehistoric Human Mobility in Polynesia Indicated by Mtdna from the Pacific Rat (Rattus Exulans͞population Mobility)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Memory Work on R ¯Ekohu (Chatham Islands) Kingsley Baird
Memory Connection Volume 3 Number 1 © 2019 The Memory Waka Hokopanopano Ka Toi Moriori (Reigniting Moriori Arts): Memory Work on R ¯ekohu (Chatham Islands) Kingsley Baird Hokopanopano Ka Toi Moriori (Reigniting Moriori Arts): Memory Work on R ¯ekohu (Chatham Islands)—Kingsley Baird Hokopanopano Ka Toi Moriori (Reigniting Moriori Arts): Memory Work on R ¯ekohu (Chatham Islands) Kingsley Baird Abstract Since European discovery of Re¯kohu (Chatham Islands) in 1791, the pacifist Moriori population declined rapidly as a result of introduced diseases (to which they had no immunity) and killing and enslavement by M¯aori iwi (tribes) from the New Zealand ‘mainland’ following their invasion in 1835. When (full-blooded) Tame Horomona Rehe—described on his headstone as the ‘last of the Morioris’— died in 1933, the Moriori were widely considered to be an extinct people. In February 2016, Moriori rangata m¯a tua (elders) and rangatehi (youth), artists and designers, archaeologists, a conservator and an arborist gathered at Ko¯ pinga Marae on Re¯kohu to participate in a w¯a nanga organized by the Hokotehi Moriori Trust. Its purpose was to enlist the combined expertise and commitment of the participants to hokopanopano ka toi Moriori (reignite Moriori arts)—principally those associated with r¯a kau momori (‘carving’ on living ko¯ pi trees)—through discussion, information exchange, speculation, toolmaking and finally, tree carving. In addition to providing a brief cultural and historical background, this paper recounts some of the memory work of the w¯a nanga from the perspective of one of the participants whose fascination for Moriori and the resilience of their culture developed from Michael King’s 1989 book, Moriori: A People Rediscovered. -

Biodiversity of the Kermadec Islands and Offshore Waters of the Kermadec Ridge: Report of a Coastal, Marine Mammal and Deep-Sea Survey (TAN1612)
Biodiversity of the Kermadec Islands and offshore waters of the Kermadec Ridge: report of a coastal, marine mammal and deep-sea survey (TAN1612) New Zealand Aquatic Environment and Biodiversity Report No. 179 Clark, M.R.; Trnski, T.; Constantine, R.; Aguirre, J.D.; Barker, J.; Betty, E.; Bowden, D.A.; Connell, A.; Duffy, C.; George, S.; Hannam, S.; Liggins, L..; Middleton, C.; Mills, S.; Pallentin, A.; Riekkola, L.; Sampey, A.; Sewell, M.; Spong, K.; Stewart, A.; Stewart, R.; Struthers, C.; van Oosterom, L. ISSN 1179-6480 (online) ISSN 1176-9440 (print) ISBN 978-1-77665-481-9 (online) ISBN 978-1-77665-482-6 (print) January 2017 Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries websites at: http://www.mpi.govt.nz/news-resources/publications.aspx http://fs.fish.govt.nz go to Document library/Research reports © Crown Copyright - Ministry for Primary Industries TABLE OF CONTENTS EXECUTIVE SUMMARY 1 1. INTRODUCTION 3 1.1 Objectives: 3 1.2 Objective 1: Benthic offshore biodiversity 3 1.3 Objective 2: Marine mammal research 4 1.4 Objective 3: Coastal biodiversity and connectivity 5 2. METHODS 5 2.1 Survey area 5 2.2 Survey design 6 Offshore Biodiversity 6 Marine mammal sampling 8 Coastal survey 8 Station recording 8 2.3 Sampling operations 8 Multibeam mapping 8 Photographic transect survey 9 Fish and Invertebrate sampling 9 Plankton sampling 11 Catch processing 11 Environmental sampling 12 Marine mammal sampling 12 Dive sampling operations 12 Outreach 13 3. -

Rangi Above/Papa Below, Tangaroa Ascendant, Water All Around Us: Austronesian Creation Myths
UNLV Retrospective Theses & Dissertations 1-1-2005 Rangi above/Papa below, Tangaroa ascendant, water all around us: Austronesian creation myths Amy M Green University of Nevada, Las Vegas Follow this and additional works at: https://digitalscholarship.unlv.edu/rtds Repository Citation Green, Amy M, "Rangi above/Papa below, Tangaroa ascendant, water all around us: Austronesian creation myths" (2005). UNLV Retrospective Theses & Dissertations. 1938. http://dx.doi.org/10.25669/b2px-g53a This Thesis is protected by copyright and/or related rights. It has been brought to you by Digital Scholarship@UNLV with permission from the rights-holder(s). You are free to use this Thesis in any way that is permitted by the copyright and related rights legislation that applies to your use. For other uses you need to obtain permission from the rights-holder(s) directly, unless additional rights are indicated by a Creative Commons license in the record and/ or on the work itself. This Thesis has been accepted for inclusion in UNLV Retrospective Theses & Dissertations by an authorized administrator of Digital Scholarship@UNLV. For more information, please contact [email protected]. RANGI ABOVE/ PAPA BELOW, TANGAROA ASCENDANT, WATER ALL AROUND US: AUSTRONESIAN CREATION MYTHS By Amy M. Green Bachelor of Arts University of Nevada, Las Vegas 2004 A thesis submitted in partial fulfillment of the requirements for the Master of Arts Degree in English Department of English College of Liberal Arts Graduate College University of Nevada, Las Vegas May 2006 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. UMI Number: 1436751 Copyright 2006 by Green, Amy M. -

Hawaiki Cable Project Presentation
South Pacific region specificity L.os Angeles Hawaii q Huge distances Hawaii q Limited populaons Guam Kiribati Nauru q Isolaon issues Tuvalu Tokelau Papua New Guinea Solomon Wallis Samo a American Samoa q Need for cheaper Vanuatu French Polynesia and faster bandwidth New CaledoniaFiji Niue Tong Cook Island a q Satellite bandwidth Norfolk Sydney price over 1500 USD / Mbps Auckland 2 Existing systems in South Pacific region q Southern Cross : Sydney - Auckland - Hawaii - US west coast - Suva - Sydney ü Capacity: 6 Tb/s ü End of life: 2020 q Endeavour (Telstra) : Sydney - Hawaii HawaiiHawaii ü Capacity: 1,2 Tb/s ü End of life: 2034 Guam q Gondwana : Nouméa - Sydney ü Capacity: 640 Gb/s Madang Honiara Apia ü End of life: 2033 Wallis Port Vila Pago Pago Tahiti Suva q Honotua : Tahi - Hawaii Noumea Nuku’alofa ü Capacity: 640 Gb/s Norfolk Is. ü End of life: 2035 Sydney Auckland q ASH : Pago-Pago - Hawaii ü Capacity: 1 Gb/s ü End of life: 2014 / 2015 ? (no more spare parts) ü SAS cable : Apia - Pago Pago 3 Hawaiki cable project overview q Project summary ü Provide internaonal bandwidth to Australia + New Zealand + Pacific Islands ü Propose point to point capacity via 100 Gb/s wavelengths ü System design capacity : 20 Tbps ü 2 step project q Time schedule ü Q1 2013 : signature of supplier contract ü Service date : 2015 q Project development by Intelia (www.intelia.nc) ü Leading telecom integrator ü Partnership with Ericsson, ZTE, Telstra, Prysmian, etc… ü 2011 turnover > USD 40M Commercial references : ü Supply and installaon of 3G+ mobile network in NC ü IP transit service for Gondwana cable in Sydney Submarine cable experience - in partnership with ASN: ü New Caledonia cable : Gondwana in 2008 - 2 100 km ü French Polynesia cable : Honotua in 2010 - 4 500 km 4 Hawaiki Cable Step 1 Main backbone / Strategic route Hawaii California Hawaii Guam Madang Honiara Pago Pago Wallis Apia Tahiti Port Vila Suva Noumea Niue Nuku’alofa Rarotonga Norfolk Is. -

Assessment of the Weed Control Programme on Raoul Island, Kermadec Group
4000 in extent (Devine 1975).At Low Flat shore hibiscus is extensive in the south-western corner of the flat. In 1993 one small plant was found growing above the strand line at Coral Bay and the same plant was first seen in 1991 (Clapham 1991b). This plant may have established from seed as shore hibiscus is a common strand plant in the Pacific (Merrill 1940). What is uncertain, though, is where the seed originated from. Seedlings have only occasionally been recorded under the large stands on Raoul Island (Clapham 1991b), and seed set has not been observed. It is possible that the Coral Bay plant germinated from seed dispersed from elsewhere in the Pacific. Alternatively, the plant at Coral Bay could have established from a stem fragment washed around the coast from Denham Bay or Low Flat. However, given that all known stands are some distance from the sea, this explanation is less likely. At the start of the weed eradication programme, shore hibiscus was listed as a category A plant (Devine 1977). In 1980, Sykes noted that the plants at Low Flat and Denham Bay had not increased much and because they were only slightly increasing through vegetative layering should be accorded low priority in the eradication programme. 7.3.2 Ecology Shore hibiscus is a sprawling shrub up to 4 m tall belonging to the mallow family (Malvaceae). Leaves are densely hairy below and velvety to touch, almost circular and c. 10-30 cm diam. Yellow flowers with dark purple centres are c. 30-70 mm long. -

A Virtual Reality Simulation for Non
KILO HŌKŪ: A VIRTUAL REALITY SIMULATION FOR NON-INSTRUMENT HAWAIIAN NAVIGATION A THESIS SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAWAIʻI AT MĀNOA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE IN COMPUTER SCIENCE DECEMBER 2019 By Patrick A. Karjala Thesis Committee: Jason Leigh, Chairperson Scott Robertson Philip Johnson Keywords: virtual reality, wayfinding, education, Polynesian voyaging, Hōkūleʻa Acknowledgements Special thanks are given to the Kilo Hōkū development team, Dean Lodes, Kari Noe, and Anna Sikkink; the Laboratory for Advanced Visualization and Applications (LAVA) at the University of Hawaiʻi at Mānoa; the ʻImiloa Astronomy Center (IAC); and the Polynesian Voyaging Society (PVS). In particular, thanks are given to Kaʻiulani Murphy (PVS) and her University of Hawaiʻi at Mānoa Fall 2016 Wayfinding students; to Miki Tomita (PVS); and to Celeste Haʻo (IAC) for feedback on the initial prototype of the simulation. Additional thanks are given to the instructors of the Fall 2018 sections of Hawaiian Studies 281, Kaʻiulani Murphy and Ian Kekaimalu Lee, for their feedback and time with their classes for the study, and to the Honolulu Community College Marine Education Technical Center, the Polynesian Voyaging Society, and the Leeward Community College Wai‘anae Moku campus staff for assistance with space use and coordination. Funding for this project was made possible by Chris Lee of the Academy for Creative Media System at the University of Hawaiʻi at Mānoa. The name Hōkūleʻa and the sailing vessel Hōkūleʻa are trademarks of and are owned by the Polynesian Voyaging Society, and are used within the Kilo Hōkū simulation with permission. -

My Friend, the Volcano (Adapted from the 2004 Submarine Ring of Fire Expedition)
New Zealand American Submarine Ring of Fire 2007 My Friend, The Volcano (adapted from the 2004 Submarine Ring of Fire Expedition) FOCUS MAXIMUM NUMBER OF STUDENTS Ecological impacts of volcanism in the Mariana 30 and Kermadec Islands KEY WORDS GRADE LEVEL Ring of Fire 5-6 (Life Science/Earth Science) Asthenosphere Lithosphere FOCUS QUESTION Magma What are the ecological impacts of volcanic erup- Fault tions on tropical island arcs? Transform boundary Convergent boundary LEARNING OBJECTIVES Divergent boundary Students will be able to describe at least three Subduction beneficial impacts of volcanic activity on marine Tectonic plate ecosystems. BACKGROUND INFORMATION Students will be able to explain the overall tec- The Submarine Ring of Fire is an arc of active vol- tonic processes that cause volcanic activity along canoes that partially encircles the Pacific Ocean the Mariana Arc and Kermadec Arc. Basin, including the Kermadec and Mariana Islands in the western Pacific, the Aleutian Islands MATERIALS between the Pacific and Bering Sea, the Cascade Copies of “Marianas eruption killed Anatahan’s Mountains in western North America, and numer- corals,” one copy per student or student group ous volcanoes on the western coasts of Central (from http://www.cdnn.info/eco/e030920/e030920.html) America and South America. These volcanoes result from the motion of large pieces of the AUDIO/VISUAL MATERIALS Earth’s crust known as tectonic plates. None Tectonic plates are portions of the Earth’s outer TEACHING TIME crust (the lithosphere) about 5 km thick, as Two or three 45-minute class periods, plus time well as the upper 60 - 75 km of the underlying for student research mantle. -

Obsidian from Macauley Island: a New Zealand Connection
www.aucklandmuseum.com Obsidian from Macauley Island: a New Zealand connection Louise Furey Auckland War Memorial Museum Callan Ross-Sheppard McGill University, Montreal Kath E. Prickett Auckland War Memorial Museum Abstract An obsidian flake collected from Macauley Island during the Kermadec Expedition has been analysed to determine the source. The result indicates a Mayor Island, New Zealand, origin, supporting previous results on obsidian from Raoul Island that Polynesians travelled back into the Pacific from New Zealand. Keywords obsidian; Pacific colonisation; Pacific voyaging; Macauley Island. INTRODUCTION The steep terrain meant there were only a few suitable habitation places confined to flat areas of limited size on The Kermadec Islands comprise Raoul Island, the largest the south and north coasts. The only landing places were of the group, the nearby Meyer Islands and Herald adjacent to these flats. Botanical surveys have recorded Islets, and 100–155 km to the south are Macauley, the presence of Polynesian tropical cultigens including Curtis and Cheeseman iIslands, and L’Esperence Rock. candlenut (Aleurites monuccana) and ti (Cordyline Raoul is approximately 1000 km to the northeast of fruticosa) (Sykes 1977) which are not endemic to the New Zealand. During the 2011 Kermadec Expedition, island and it is assumed they were transported there by samples of naturally occurring obsidian were obtained Polynesian settlers. Polynesian tuberous vegetables such from Raoul, the Meyer Islands, and from the sea floor off as taro (Colocasia esculenta) and kumara (Ipomoea Curtis Island. These are now in the Auckland Museum batatas) were also recorded but their introduction collection. More importantly however was the single attributed to later 19th century immigrants. -

Rekohu Report (2016 Newc).Vp
Rekohu REKOHU AReporton MorioriandNgatiMutungaClaims in the Chatham Islands Wa i 6 4 WaitangiTribunalReport2001 The cover design by Cliff Whiting invokes the signing of the Treaty of Waitangi and the consequent interwoven development of Maori and Pakeha history in New Zealand as it continuously unfoldsinapatternnotyetcompletelyknown AWaitangiTribunalreport isbn 978-1-86956-260-1 © Waitangi Tribunal 2001 Reprinted with corrections 2016 www.waitangi-tribunal.govt.nz Produced by the Waitangi Tribunal Published by Legislation Direct, Wellington, New Zealand Printed by Printlink, Lower Hutt, New Zealand Set in Adobe Minion and Cronos multiple master typefaces e nga mana,e nga reo,e nga karangaranga maha tae noa ki nga Minita o te Karauna. ko tenei te honore,hei tuku atu nga moemoea o ratou i kawea te kaupapa nei. huri noa ki a ratou kua wheturangitia ratou te hunga tautoko i kokiri,i mau ki te kaupapa,mai te timatanga,tae noa ki te puawaitanga o tenei ripoata. ahakoa kaore ano ki a kite ka tangi,ka mihi,ka poroporoakitia ki a ratou. ki era o nga totara o Te-Wao-nui-a-Tane,ki a Te Makarini,ki a Horomona ma ki a koutou kua huri ki tua o te arai haere,haere,haere haere i runga i te aroha,me nga roimata o matou kua mahue nei. e kore koutou e warewaretia. ma te Atua koutou e manaaki,e tiaki ka huri Contents Letter of Transmittal _____________________________________________________xiii 1. Summary 1.1 Background ________________________________________________________1 1.2 Historical Claims ____________________________________________________4 1.3 Contemporary Claims ________________________________________________9 1.4 Preliminary Claims __________________________________________________11 1.5 Rekohu, the Chatham Islands, or Wharekauri? _____________________________12 1.6 Concluding Remarks ________________________________________________13 2. -

Reasons to Visit The
Reasons to visit the First place in the world to greet the new dawn Home of unique Chatham Islands birds and plants Visit significant sites of history and heritage Learn about the ancient Moriori covenant of peace Go fishing and hunting Enjoy rugged and awe inspiring landscapes Meet the people of the Chatham Islands Top 20 “Must See” Attractions Admiral Gardens & Pitt Island Kahukura Studio Point Munning Seal Colony Awatotara Bush Coastal Walking Track Port Hutt Basalt Columns Stone Cottage Chatham Cottage Crafts Sunderland Flying Boat Chatham Island Food Co. Taiko Camp and Gap Sanctuary Chatham Islands Museum Tommy Solomon Eva-Cherie Artz & Memorial Statue Studio 44°s Waitangi West Fishing Charters Wharekauri Station and Kaingaroa Splatter Rock chathamislands.co.nz Kopinga Marae DOC Walks Splatter/Taniwha Rock Skirmish Bay Stay " Wharekauri The Landing Ponga Whare Sunderland Flying Boat Maunganui Stone Cottage Ocean Mail Scenic Reserve Waitangi West " Point Munning " N KAIWHATA RD Conservation Covenant O Kaingaroa PORT R Seal Colony HU T Nikau Bush Port TT RD H R Conservation Area Te Whakaru Graveyard Hutt D " German Missionaries Settlement Basalt Columns Go Wild Nursery Thomas Currell J M Barker (Hapupu) Port Hutt Bay Stays Admiral Garden & Kahukura Studio AIRBASE RD National Historic Reserve Henga Lodge Te Whanga Henga Scenic Reserve Lagoon Chatham Island (R kohu / Wh arekauri) Tikitiki Hill Conservation Area " Chatham Island Charters Te One Pitt Island is Lake Pitt Island Guided Access Only " Huro " Te Matarae Kopinga Marae (Rangihaute/Rangiauria) -
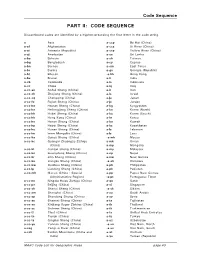
Code Sequence
Code Sequence PART II: CODE SEQUENCE Discontinued codes are identified by a hyphen preceding the first letter in the code string. a Asia a-ccp Bo Hai (China) a-af Afghanistan a-ccs Xi River (China) a-ai Armenia (Republic) a-ccy Yellow River (China) a-aj Azerbaijan a-ce Sri Lanka a-ba Bahrain a-ch Taiwan a-bg Bangladesh a-cy Cyprus a-bn Borneo a-em East Timor a-br Burma a-gs Georgia (Republic) a-bt Bhutan -a-hk Hong Kong a-bx Brunei a-ii India a-cb Cambodia a-io Indonesia a-cc China a-iq Iraq a-cc-an Anhui Sheng (China) a-ir Iran a-cc-ch Zhejiang Sheng (China) a-is Israel a-cc-cq Chongqing (China) a-ja Japan a-cc-fu Fujian Sheng (China) a-jo Jordan a-cc-ha Hainan Sheng (China) a-kg Kyrgyzstan a-cc-he Heilongjiang Sheng (China) a-kn Korea (North) a-cc-hh Hubei Sheng (China) a-ko Korea (South) a-cc-hk Hong Kong (China) a-kr Korea a-cc-ho Henan Sheng (China) a-ku Kuwait a-cc-hp Hebei Sheng (China) a-kz Kazakhstan a-cc-hu Hunan Sheng (China) a-le Lebanon a-cc-im Inner Mongolia (China) a-ls Laos a-cc-ka Gansu Sheng (China) -a-mh Macao a-cc-kc Guangxi Zhuangzu Zizhiqu a-mk Oman (China) a-mp Mongolia a-cc-ki Jiangxi Sheng (China) a-my Malaysia a-cc-kn Guangdong Sheng (China) a-np Nepal a-cc-kr Jilin Sheng (China) a-nw New Guinea a-cc-ku Jiangsu Sheng (China) -a-ok Okinawa a-cc-kw Guizhou Sheng (China) a-ph Philippines a-cc-lp Liaoning Sheng (China) a-pk Pakistan a-cc-mh Macao (China : Special a-pp Papua New Guinea Administrative Region) -a-pt Portuguese Timor a-cc-nn Ningxia Huizu Zizhiqu (China) a-qa Qatar a-cc-pe Beijing (China) a-si Singapore -

Conservation Status of New Zealand Marine Mammals, 2019 Report
2019 NEW ZEALAND THREAT CLASSIFICATION SERIES 29 Conservation status of New Zealand marine mammals, 2019 C.S. Baker, L. Boren, S. Childerhouse, R. Constantine, A. van Helden, D. Lundquist, W. Rayment and J.R. Rolfe Dedication This report is dedicated to the memory of Dr Rob Mattlin, a founding member of this Committee, who passed away in May 2019. Rob’s knowledge, insight and personality will be greatly missed by the Committee and by the marine mammals for which he was so passionate. Rob made New Zealand his home after leaving his native USA to undertake a PhD on New Zealand fur seals at Canterbury University. He proceeded to dedicate his life to marine science with a particular focus on marine mammals and spent most of his time based in New Zealand, apart from a role at the US Marine Mammal Commission. His contribution to our understanding of marine mammal biology and ecology and, in particular, his expertise in interactions with fisheries, was a key factor in achieving many positive outcomes for marine mammals in New Zealand. He made significant contributions to the conservation and management of both New Zealand sea lions and Hector’s dolphins. The Committee would like to recognise the unique balance of wisdom, humour, and tenacity that Rob brought to his work and hope that we can continue to match his enthusiasm and passion. Cover: The southern right whale (Eubalaena australis) population is recovering from perilously low numbers and is now assessed as At Risk – Recovering, an improvement on its previous Threatened status. Photo: Will Rayment.