Cryptic Species Delimitation in the Southern Appalachian Antrodiaetus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biogeography of the Caribbean Cyrtognatha Spiders Klemen Čandek1,6,7, Ingi Agnarsson2,4, Greta J
www.nature.com/scientificreports OPEN Biogeography of the Caribbean Cyrtognatha spiders Klemen Čandek1,6,7, Ingi Agnarsson2,4, Greta J. Binford3 & Matjaž Kuntner 1,4,5,6 Island systems provide excellent arenas to test evolutionary hypotheses pertaining to gene fow and Received: 23 July 2018 diversifcation of dispersal-limited organisms. Here we focus on an orbweaver spider genus Cyrtognatha Accepted: 1 November 2018 (Tetragnathidae) from the Caribbean, with the aims to reconstruct its evolutionary history, examine Published: xx xx xxxx its biogeographic history in the archipelago, and to estimate the timing and route of Caribbean colonization. Specifcally, we test if Cyrtognatha biogeographic history is consistent with an ancient vicariant scenario (the GAARlandia landbridge hypothesis) or overwater dispersal. We reconstructed a species level phylogeny based on one mitochondrial (COI) and one nuclear (28S) marker. We then used this topology to constrain a time-calibrated mtDNA phylogeny, for subsequent biogeographical analyses in BioGeoBEARS of over 100 originally sampled Cyrtognatha individuals, using models with and without a founder event parameter. Our results suggest a radiation of Caribbean Cyrtognatha, containing 11 to 14 species that are exclusively single island endemics. Although biogeographic reconstructions cannot refute a vicariant origin of the Caribbean clade, possibly an artifact of sparse outgroup availability, they indicate timing of colonization that is much too recent for GAARlandia to have played a role. Instead, an overwater colonization to the Caribbean in mid-Miocene better explains the data. From Hispaniola, Cyrtognatha subsequently dispersed to, and diversifed on, the other islands of the Greater, and Lesser Antilles. Within the constraints of our island system and data, a model that omits the founder event parameter from biogeographic analysis is less suitable than the equivalent model with a founder event. -

Final Project Completion Report
CEPF SMALL GRANT FINAL PROJECT COMPLETION REPORT Organization Legal Name: - Tarantula (Araneae: Theraphosidae) spider diversity, distribution and habitat-use: A study on Protected Area adequacy and Project Title: conservation planning at a landscape level in the Western Ghats of Uttara Kannada district, Karnataka Date of Report: 18 August 2011 Dr. Manju Siliwal Wildlife Information Liaison Development Society Report Author and Contact 9-A, Lal Bahadur Colony, Near Bharathi Colony Information Peelamedu Coimbatore 641004 Tamil Nadu, India CEPF Region: The Western Ghats Region (Sahyadri-Konkan and Malnad-Kodugu Corridors). 2. Strategic Direction: To improve the conservation of globally threatened species of the Western Ghats through systematic conservation planning and action. The present project aimed to improve the conservation status of two globally threatened (Molur et al. 2008b, Siliwal et al., 2008b) ground dwelling theraphosid species, Thrigmopoeus insignis and T. truculentus endemic to the Western Ghats through systematic conservation planning and action. Investment Priority 2.1 Monitor and assess the conservation status of globally threatened species with an emphasis on lesser-known organisms such as reptiles and fish. The present project was focused on an ignored or lesser-known group of spiders called Tarantulas/ Theraphosid spiders and provided valuable information on population status and potential conservation sites in Uttara Kannada district, which will help in future monitoring and assessment of conservation status of the two globally threatened theraphosid species T. insignis and Near Threatened T. truculentus. Investment Priority 2.3. Evaluate the existing protected area network for adequate globally threatened species representation and assess effectiveness of protected area types in biodiversity conservation. -

Tarantulas in the Pacific Northwest1
WSU Puyallup REC PLS-108 Updated July 2003 Tarantulas in the Pacific Northwest1 Tarantulas (Fig. 1) in the Pacific Northwest? Well, maybe not like the hairy monsters of the tropics, but some very interesting "atypical" species do occur here. Our species belong to the family Antrodiaetidae. One of our most common spiders is the folding-door spider, Antrodiaetus pacificus (Simon). It is a fairly large species, females ranging from 11 to 13 millimeters in length, males slightly smaller. They are generally dark brown to almost black in color with the abdomen purplish brown. Males are characterized by their long legs, slim bodies, and three tergites (hardened plates) on the abdomen. Females (Fig. 2) are more robust with only one tergite. These spiders excavate burrows in the soil or in damp, rotten wood, digging with a row of spines on each chelicer, known as a ratellum. The six to ten inch deep vertical shafts are lined with silk. The webbing extends beyond ground level as a short collar of camouflaged silk. The turret’s two sides may be drawn in by the occupant, forming two "doors" which meet in the middle. At night, Antrodiaetus assumes a foraging posture with its pedipalps and first pair of legs just touching the rim of silk at the mouth of the tube. In this position, the folding door spider can readily detect an insect moving above ground. The spider will leap out of its burrow with lightning speed, seize its victim, and drop back down, like a terrorizing Jack-in-the-box. When finished with its meal, it will add the insect's dry, dismembered body to a silk-covered trash pile at the bottom of its burrow. -
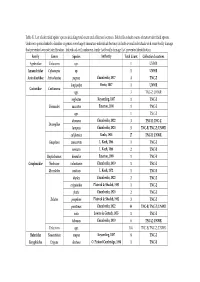
Table S1. List of Identified Spider Species Including Total Count and Collection Locations. Bold Cells Include Counts of Mature Identified Species
Table S1. List of identified spider species including total count and collection locations. Bold cells include counts of mature identified species. Unknown species linked to families or genera were largely immature individuals but may include several individuals with some bodily damage that prevented accurate identification. Individuals with unknown family had bodily damage that prevented identification. Family Genus Species Authority Total Count Collection Locations Agelenidae Unknown spp. 1 UNWR Amaurobiidae Cybaeopsis sp. 1 UNWR Antrodiaetidae Antrodiaetus pugnax Chamberlin, 1917 4 TNC-Z longipalpa Hentz, 1847 1 UNWR Corinnidae Castianeira spp. 3 TNC-Z; UNWR neglectus Keyserling, 1887 1 TNC-Z Drassodes saccatus Emerton, 1890 1 TNC-Z spp. 1 TNC-Z dromeus Chamberlin, 1922 3 TNC-B; TNC-Z Drassyllus lamprus Chamberlin, 1920 3 TNC-B; TNC-Z; UNWR californica Banks, 1904 17 TNC-B; UNWR Gnaphosa muscorum L. Koch, 1866 1 TNC-Z sericata L. Koch, 1866 2 TNC-B Haplodrassus hiemalis Emerton, 1909 1 TNC-B Gnaphosidae Nodocion voluntaries Chamberlin, 1919 1 TNC-Z Urozelotes rusticus L. Koch, 1872 1 TNC-B duplex Chamberlin, 1922 2 TNC-Z exiguioides Platnick & Shadab, 1983 1 TNC-Z fratis Chamberlin, 1920 2 TNC-Z Zelotes josephine Platnick & Shadab, 1983 3 TNC-Z puritanus Chamberlin, 1922 44 TNC-B; TNC-Z; UNWR sula Lowrie & Gertsch, 1955 1 TNC-Z tubuous Chamberlin, 1919 6 TNC-Z; UNWR Unknown spp. 184 TNC-B; TNC-Z; UNWR Hahniidae Neoantistea magna Keyserling, 1887 8 TNC-Z Linyphiidae Erigone dentosa O. Pickard-Cambridge, 1894 1 TNC-B spp. 1 TNC-Z Unknown spp. 13 TNC-Z; UNWR mccooki Montgomery, 1904 95 TNC-B; TNC-Z; UNWR Schizocosa minnesotensis Gertsch, 1934 25 TNC-B Lycosidae spp. -

Tarantulas and Social Spiders
Tarantulas and Social Spiders: A Tale of Sex and Silk by Jonathan Bull BSc (Hons) MSc ICL Thesis Presented to the Institute of Biology of The University of Nottingham in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy The University of Nottingham May 2012 DEDICATION To my parents… …because they both said to dedicate it to the other… I dedicate it to both ii ACKNOWLEDGEMENTS First and foremost I would like to thank my supervisor Dr Sara Goodacre for her guidance and support. I am also hugely endebted to Dr Keith Spriggs who became my mentor in the field of RNA and without whom my understanding of the field would have been but a fraction of what it is now. Particular thanks go to Professor John Brookfield, an expert in the field of biological statistics and data retrieval. Likewise with Dr Susan Liddell for her proteomics assistance, a truly remarkable individual on par with Professor Brookfield in being able to simplify even the most complex techniques and analyses. Finally, I would really like to thank Janet Beccaloni for her time and resources at the Natural History Museum, London, permitting me access to the collections therein; ten years on and still a delight. Finally, amongst the greats, Alexander ‘Sasha’ Kondrashov… a true inspiration. I would also like to express my gratitude to those who, although may not have directly contributed, should not be forgotten due to their continued assistance and considerate nature: Dr Chris Wade (five straight hours of help was not uncommon!), Sue Buxton (direct to my bench creepy crawlies), Sheila Keeble (ventures and cleans where others dare not), Alice Young (read/checked my thesis and overcame her arachnophobia!) and all those in the Centre for Biomolecular Sciences. -

Species Are Hypotheses: Avoid Connectivity Assessments Based on Pillars of Sand Eric Pante, Nicolas Puillandre, Amélia Viricel, Sophie Arnaud-Haond, D
Species are hypotheses: avoid connectivity assessments based on pillars of sand Eric Pante, Nicolas Puillandre, Amélia Viricel, Sophie Arnaud-Haond, D. Aurelle, Magalie Castelin, Anne Chenuil, Christophe Destombe, Didier Forcioli, Myriam Valero, et al. To cite this version: Eric Pante, Nicolas Puillandre, Amélia Viricel, Sophie Arnaud-Haond, D. Aurelle, et al.. Species are hypotheses: avoid connectivity assessments based on pillars of sand. Molecular Ecology, Wiley, 2015, 24 (3), pp.525-544. hal-02002440 HAL Id: hal-02002440 https://hal.archives-ouvertes.fr/hal-02002440 Submitted on 31 Jan 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Molecular Ecology Species are hypotheses : avoid basing connectivity assessments on pillars of sand. Journal:For Molecular Review Ecology Only Manuscript ID: Draft Manuscript Type: Invited Reviews and Syntheses Date Submitted by the Author: n/a Complete List of Authors: Pante, Eric; UMR 7266 CNRS - Université de La Rochelle, Puillandre, Nicolas; MNHN, Systematique & Evolution Viricel, Amélia; UMR 7266 CNRS - -

Species Delimitation and Phylogeography of Aphonopelma Hentzi (Araneae, Mygalomorphae, Theraphosidae): Cryptic Diversity in North American Tarantulas
Species Delimitation and Phylogeography of Aphonopelma hentzi (Araneae, Mygalomorphae, Theraphosidae): Cryptic Diversity in North American Tarantulas Chris A. Hamilton1*, Daniel R. Formanowicz2, Jason E. Bond1 1 Auburn University Museum of Natural History and Department of Biological Sciences, Auburn University, Auburn, Alabama, United States of America, 2 Department of Biology, The University of Texas at Arlington, Arlington, Texas, United States of America Abstract Background: The primary objective of this study is to reconstruct the phylogeny of the hentzi species group and sister species in the North American tarantula genus, Aphonopelma, using a set of mitochondrial DNA markers that include the animal ‘‘barcoding gene’’. An mtDNA genealogy is used to consider questions regarding species boundary delimitation and to evaluate timing of divergence to infer historical biogeographic events that played a role in shaping the present-day diversity and distribution. We aimed to identify potential refugial locations, directionality of range expansion, and test whether A. hentzi post-glacial expansion fit a predicted time frame. Methods and Findings: A Bayesian phylogenetic approach was used to analyze a 2051 base pair (bp) mtDNA data matrix comprising aligned fragments of the gene regions CO1 (1165 bp) and ND1-16S (886 bp). Multiple species delimitation techniques (DNA tree-based methods, a ‘‘barcode gap’’ using percent of pairwise sequence divergence (uncorrected p- distances), and the GMYC method) consistently recognized a number of divergent and genealogically exclusive groups. Conclusions: The use of numerous species delimitation methods, in concert, provide an effective approach to dissecting species boundaries in this spider group; as well they seem to provide strong evidence for a number of nominal, previously undiscovered, and cryptic species. -

A Stable Phylogenomic Classification of Travunioidea (Arachnida, Opiliones, Laniatores) Based on Sequence Capture of Ultraconserved Elements
A stable phylogenomic classification of Travunioidea (Arachnida, Opiliones, Laniatores) based on sequence capture of ultraconserved elements The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Derkarabetian, Shahan, James Starrett, Nobuo Tsurusaki, Darrell Ubick, Stephanie Castillo, and Marshal Hedin. 2018. “A stable phylogenomic classification of Travunioidea (Arachnida, Opiliones, Laniatores) based on sequence capture of ultraconserved elements.” ZooKeys (760): 1-36. doi:10.3897/zookeys.760.24937. http://dx.doi.org/10.3897/zookeys.760.24937. Published Version doi:10.3897/zookeys.760.24937 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:37298544 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA A peer-reviewed open-access journal ZooKeys 760: 1–36 (2018) A stable phylogenomic classification of Travunioidea... 1 doi: 10.3897/zookeys.760.24937 RESEARCH ARTICLE http://zookeys.pensoft.net Launched to accelerate biodiversity research A stable phylogenomic classification of Travunioidea (Arachnida, Opiliones, Laniatores) based on sequence capture of ultraconserved elements Shahan Derkarabetian1,2,7 , James Starrett3, Nobuo Tsurusaki4, Darrell Ubick5, Stephanie Castillo6, Marshal Hedin1 1 Department of Biology, San Diego State University, San -

Phylogenomic Analysis and Revised Classification of Atypoid Mygalomorph Spiders (Araneae, Mygalomorphae), with Notes on Arachnid Ultraconserved Element Loci
Phylogenomic analysis and revised classification of atypoid mygalomorph spiders (Araneae, Mygalomorphae), with notes on arachnid ultraconserved element loci Marshal Hedin1, Shahan Derkarabetian1,2,3, Adan Alfaro1, Martín J. Ramírez4 and Jason E. Bond5 1 Department of Biology, San Diego State University, San Diego, CA, United States of America 2 Department of Biology, University of California, Riverside, Riverside, CA, United States of America 3 Department of Organismic and Evolutionary Biology, Museum of Comparative Zoology, Harvard University, Cambridge, MA, United States of America 4 Division of Arachnology, Museo Argentino de Ciencias Naturales ``Bernardino Rivadavia'', Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Buenos Aires, Argentina 5 Department of Entomology and Nematology, University of California, Davis, CA, United States of America ABSTRACT The atypoid mygalomorphs include spiders from three described families that build a diverse array of entrance web constructs, including funnel-and-sheet webs, purse webs, trapdoors, turrets and silken collars. Molecular phylogenetic analyses have generally supported the monophyly of Atypoidea, but prior studies have not sampled all relevant taxa. Here we generated a dataset of ultraconserved element loci for all described atypoid genera, including taxa (Mecicobothrium and Hexurella) key to understanding familial monophyly, divergence times, and patterns of entrance web evolution. We show that the conserved regions of the arachnid UCE probe set target exons, such that it should be possible to combine UCE and transcriptome datasets in arachnids. We also show that different UCE probes sometimes target the same protein, and under the matching parameters used here show that UCE alignments sometimes include non- Submitted 1 February 2019 orthologs. Using multiple curated phylogenomic matrices we recover a monophyletic Accepted 28 March 2019 Published 3 May 2019 Atypoidea, and reveal that the family Mecicobothriidae comprises four separate and divergent lineages. -

Scope: Munis Entomology & Zoology Publishes a Wide Variety of Papers
_____________ Mun. Ent. Zool. Vol. 2, No. 1, January 2007___________ I MUNIS ENTOMOLOGY & ZOOLOGY Ankara / Turkey II _____________ Mun. Ent. Zool. Vol. 2, No. 1, January 2007___________ Scope: Munis Entomology & Zoology publishes a wide variety of papers on all aspects of Entomology and Zoology from all of the world, including mainly studies on systematics, taxonomy, nomenclature, fauna, biogeography, biodiversity, ecology, morphology, behavior, conservation, pa!eobiology and other aspects are appropriate topics for papers submitted to Munis Entomology & Zoology. Submission of Manuscripts: Works published or under consideration elsewhere (including on the internet) will not be accepted. At first submission, one double spaced hard copy (text and tables) with figures (may not be original) must be sent to the Editors, Dr. Hüseyin Özdikmen for publication in MEZ. All manuscripts should be submitted as Word file or PDF file in an e-mail attachment. If electronic submission is not possible due to limitations of electronic space at the sending or receiving ends, unavailability of e-mail, etc., we will accept ―hard‖ versions, in triplicate, accompanied by an electronic version stored in a floppy disk, a CD-ROM. Review Process: When submitting manuscripts, all authors provides the name, of at least three qualified experts (they also provide their address, subject fields and e-mails). Then, the editors send to experts to review the papers. The review process should normally be completed within 45-60 days. After reviewing papers by reviwers: Rejected papers are discarded. For accepted papers, authors are asked to modify their papers according to suggestions of the reviewers and editors. Final versions of manuscripts and figures are needed in a digital format. -

Folding Door Spider
Folding Door Spider Order: Arachnida Family: Antrodiaetidae Genus/species: Antrodiaetus species Description: The folding door spider is tan to chestnut brown about ¼” – 1 ½ “long. They have rakelike rows of spines on each jaw, or chelicera, which they use for digging. They have 3 claws on each foot. Folding door spiders have 1-4 small crescent shaped sclerites (hardened plates) on the top of the abdomen. They have 6 eyes in two groups of three. Habitat: Folding door spiders excavate burrows in the ground that they line with silk. At the entrance they construct a collapsible “trap door”. When active they open their trapdoor at night and wait just inside the entrance to strike at passing prey. The spider detects the prey by sensing vibrations. When mature, males abandon their burrows during rainy periods and wander in search of females. Life Cycle: Most spiders live 1 to 2 years. The adult male begins courtship after his palp is filled with sperm and he has found a female. Soon after mating the male dies. After a week or more the mated female deposits her eggs in a silken sac. Weeks later, or sometimes not until the following spring, the young spiderlings emerge. Economic Importance & Management: All spiders are voracious predators. Folding Door spiders feed on insects, and are considered highly beneficial because they keep the burgeoning insect populations in check. References: The Audubon Society Field Guide to NA Lorus & Marjery Milne Spiders! Usually helpful, sometimes a nuisance, Jean Natter Spiders of NA-identification manual, D. Ubick, P. Paquin P.E .Cushing and V. -

Multilocus Species Delimitation in a Complex of Morphologically Conserved Trapdoor Spiders (Mygalomorphae, Antrodiaetidae, Aliatypus)
Syst. Biol. 62(6):805–823, 2013 © The Author(s) 2013. Published by Oxford University Press, on behalf of the Society of Systematic Biologists. All rights reserved. For Permissions, please email: [email protected] DOI:10.1093/sysbio/syt041 Advance Access publication June 13, 2013 Multilocus Species Delimitation in a Complex of Morphologically Conserved Trapdoor Spiders (Mygalomorphae, Antrodiaetidae, Aliatypus) ,∗ JORDAN D. SATLER1 ,BRYAN C. CARSTENS1, AND MARSHAL HEDIN2 1Department of Evolution, Ecology and Organismal Biology, The Ohio University, Columbus, OH 43210, USA and 2Department of Biology, San Diego University, San Diego, CA 92182, USA ∗ Correspondence to be sent to: Department of Evolution, Ecology and Organismal Biology, 318 West 12th Avenue, The Ohio State University, Columbus, OH 43210, USA; E-mail: [email protected] Received 17 December 2012; reviews returned 4 March 2013; accepted 7 June 2013 Associate Editor: Robb Brumfield Abstract.—Species are a fundamental unit for biological studies, yet no uniform guidelines exist for determining species limits in an objective manner. Given the large number of species concepts available, defining species can be both highly Downloaded from subjective and biased. Although morphology has been commonly used to determine species boundaries, the availability and prevalence of genetic data has allowed researchers to use such data to make inferences regarding species limits. Genetic data also have been used in the detection of cryptic species, where other lines of evidence (morphology in particular) may underestimate species diversity. In this study, we investigate species limits in a complex of morphologically conserved trapdoor spiders (Mygalomorphae, Antrodiaetidae, Aliatypus) from California. Multiple approaches were used to determine species boundaries in this highly genetically fragmented group, including both multilocus discovery and validation http://sysbio.oxfordjournals.org/ approaches (plus a chimeric approach).