Phylogenomic Analysis and Revised Classification of Atypoid Mygalomorph Spiders (Araneae, Mygalomorphae), with Notes on Arachnid Ultraconserved Element Loci
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biogeography of the Caribbean Cyrtognatha Spiders Klemen Čandek1,6,7, Ingi Agnarsson2,4, Greta J
www.nature.com/scientificreports OPEN Biogeography of the Caribbean Cyrtognatha spiders Klemen Čandek1,6,7, Ingi Agnarsson2,4, Greta J. Binford3 & Matjaž Kuntner 1,4,5,6 Island systems provide excellent arenas to test evolutionary hypotheses pertaining to gene fow and Received: 23 July 2018 diversifcation of dispersal-limited organisms. Here we focus on an orbweaver spider genus Cyrtognatha Accepted: 1 November 2018 (Tetragnathidae) from the Caribbean, with the aims to reconstruct its evolutionary history, examine Published: xx xx xxxx its biogeographic history in the archipelago, and to estimate the timing and route of Caribbean colonization. Specifcally, we test if Cyrtognatha biogeographic history is consistent with an ancient vicariant scenario (the GAARlandia landbridge hypothesis) or overwater dispersal. We reconstructed a species level phylogeny based on one mitochondrial (COI) and one nuclear (28S) marker. We then used this topology to constrain a time-calibrated mtDNA phylogeny, for subsequent biogeographical analyses in BioGeoBEARS of over 100 originally sampled Cyrtognatha individuals, using models with and without a founder event parameter. Our results suggest a radiation of Caribbean Cyrtognatha, containing 11 to 14 species that are exclusively single island endemics. Although biogeographic reconstructions cannot refute a vicariant origin of the Caribbean clade, possibly an artifact of sparse outgroup availability, they indicate timing of colonization that is much too recent for GAARlandia to have played a role. Instead, an overwater colonization to the Caribbean in mid-Miocene better explains the data. From Hispaniola, Cyrtognatha subsequently dispersed to, and diversifed on, the other islands of the Greater, and Lesser Antilles. Within the constraints of our island system and data, a model that omits the founder event parameter from biogeographic analysis is less suitable than the equivalent model with a founder event. -

Cryptic Species Delimitation in the Southern Appalachian Antrodiaetus
Cryptic species delimitation in the southern Appalachian Antrodiaetus unicolor (Araneae: Antrodiaetidae) species complex using a 3RAD approach Lacie Newton1, James Starrett1, Brent Hendrixson2, Shahan Derkarabetian3, and Jason Bond4 1University of California Davis 2Millsaps College 3Harvard University 4UC Davis May 5, 2020 Abstract Although species delimitation can be highly contentious, the development of reliable methods to accurately ascertain species boundaries is an imperative step in cataloguing and describing Earth's quickly disappearing biodiversity. Spider species delimi- tation remains largely based on morphological characters; however, many mygalomorph spider populations are morphologically indistinguishable from each other yet have considerable molecular divergence. The focus of our study, Antrodiaetus unicolor species complex which contains two sympatric species, exhibits this pattern of relative morphological stasis with considerable genetic divergence across its distribution. A past study using two molecular markers, COI and 28S, revealed that A. unicolor is paraphyletic with respect to A. microunicolor. To better investigate species boundaries in the complex, we implement the cohesion species concept and employ multiple lines of evidence for testing genetic exchangeability and ecological interchange- ability. Our integrative approach includes extensively sampling homologous loci across the genome using a RADseq approach (3RAD), assessing population structure across their geographic range using multiple genetic clustering analyses that include STRUCTURE, PCA, and a recently developed unsupervised machine learning approach (Variational Autoencoder). We eval- uate ecological similarity by using large-scale ecological data for niche-based distribution modeling. Based on our analyses, we conclude that this complex has at least one additional species as well as confirm species delimitations based on previous less comprehensive approaches. -

Arachnida: Araneae) Para México Y Listado Actualizado De La Araneofauna Del Estado De Coahuila
ISSN 0065-1737 (NUEVA SERIE) 34(1) 2018 e ISSN 2448-8445 NUEVOS REGISTROS DE ARAÑAS (ARACHNIDA: ARANEAE) PARA MÉXICO Y LISTADO ACTUALIZADO DE LA ARANEOFAUNA DEL ESTADO DE COAHUILA NEW RECORDS OF SPIDERS (ARACHNIDA: ARANEAE) FROM MEXICO AND LISTING OF SPIDERS FROM COAHUILA STATE Marco Antonio DESALES-LARA,1,2 María Luisa JIMÉNEZ3,* y Pablo CORCUERA2 1 Doctorado en Ciencias Biológicas y de la Salud, Universidad Autónoma Metropolitana-Iztapalapa (UAM-I). Av. San Rafael Atlixco 186. Col. Vicentina Iztapalapa, C.P. 09340, Cd de México, México <[email protected]> 2 Laboratorio de Ecología Animal, Departamento de Biología, Universidad Autónoma Metropolitana-Iztapalapa (UAM-I). Av. San Rafael Atlixco 186. Col. Vicentina Iztapalapa, C.P. 09340, Cd de México, México. <pcmr@ xanum.uam.mx> 3 Laboratorio de Aracnología y Entomología, Centro de Investigaciones Biológicas del Noroeste. Instituto Politécnico Nacional 195, Col. Playa Palo de Santa Rita Sur, C.P. 23096 La Paz, Baja California Sur, México. <[email protected]> *Autor para correspondencia: <[email protected]> Recibido: 27/06/2017; aceptado: 01/12/2017 Editor responsable: Guillermo Ibarra Núñez Desales-Lara, M. A., Jiménez, M. L. y Corcuera, P. (2018) Nuevos Desales-Lara, M. A., Jiménez, M. L., & Corcuera, P. (2018) New registros de arañas (Arachnida: Araneae) para México y listado records of spiders (Arachnida: Araneae) from Mexico and listing actualizado de la araneofauna del estado de Coahuila. Acta Zooló- of spiders from Coahuila state. Acta Zoológica Mexicana (n.s), gica Mexicana (n.s), 34(1), 50-63. 34(1), 50-63. RESUMEN. Se dan a conocer cuatro nuevos registros de especies ABSTRACT. -

Araneae, Dipluridae) Groups from Basal to Distal; Groups Are Divided by Hyphens
Bull. Br. arachnol. Soc. (2009) 14 (9), 365–367 365 A new genus and species of the subfamily number ( ). If any data are given in brackets, they reflect Diplurinae (Araneae, Dipluridae) groups from basal to distal; groups are divided by hyphens. Abbreviations: AME=anterior median eyes, Bastian Drolshagen ALE=anterior lateral eyes, PME=posterior median Institute of Environmental Sciences, eyes, PLE=posterior lateral eyes, Ta=tarsus, Mt= University of Koblenz-Landau, metatarsus, Ti=tibia, Pa=patella, Fe=femur, Fortstrasse 7, 76829 Landau, Germany Tr=trochanter, Co=coxa, Ma=maxillae, PMS= posterior median spinnerets, PLS=posterior lateral and spinnerets, l=lateral, pl=prolateral, rl=retrolateral, v=ventral, d=dorsal, juv=juvenile, STC=superior tar- Christian M. Bäckstam sal claw. Lindevägen 40, According to the Brazilian regulations for loaned S-120 48 Enskede gård, Sweden material, with the approval of the curator of the arach- nid collection of the SMNK, Dr Hubert Höfer, Summary the holotype and the juvenile male paratype will be A new genus and species of the subfamily Diplurinae is deposited at INPA. Abbreviations of institutes and described, Metriura striatipes gen. et sp. nov. The genus museums: SMNK=Staatliches Museum für Naturkunde differs from other diplurine genera by the presence of erect Karlsruhe, ZMB=Museum für Naturkunde Berlin, setae in front of the fovea, a large labiosternal suture and INPA=Instituto Nacional de Pesquisas do Amazônia. the strongly curved base of metatarsus I in males. Examined (type-) material: Linothele curvitarsis Karsch, 1879, holotype juv. _, ZMB Arach-458 and Arach-458a (tarsal claw preparation), Caracas, Introduction Venezuela. During examination of supposed Linothele Karsch, 1879 material at the Staatliches Museum für Metriura gen. -

Transcriptome Characterization of the Aptostichus Atomarius Species Complex Nicole L
Garrison et al. BMC Evolutionary Biology (2020) 20:68 https://doi.org/10.1186/s12862-020-01606-7 RESEARCH ARTICLE Open Access Shifting evolutionary sands: transcriptome characterization of the Aptostichus atomarius species complex Nicole L. Garrison1*, Michael S. Brewer2 and Jason E. Bond3 Abstract Background: Mygalomorph spiders represent a diverse, yet understudied lineage for which genomic level data has only recently become accessible through high-throughput genomic and transcriptomic sequencing methods. The Aptostichus atomarius species complex (family Euctenizidae) includes two coastal dune endemic members, each with inland sister species – affording exploration of dune adaptation associated patterns at the transcriptomic level. We apply an RNAseq approach to examine gene family conservation across the species complex and test for patterns of positive selection along branches leading to dune endemic species. Results: An average of ~ 44,000 contigs were assembled for eight spiders representing dune (n = 2), inland (n = 4), and atomarius species complex outgroup taxa (n = 2). Transcriptomes were estimated to be 64% complete on average with 77 spider reference orthologs missing from all taxa. Over 18,000 orthologous gene clusters were identified within the atomarius complex members, > 5000 were detected in all species, and ~ 4700 were shared between species complex members and outgroup Aptostichus species. Gene family analysis with the FUSTr pipeline identified 47 gene families appearing to be under selection in the atomarius ingroup; four of the five top clusters include sequences strongly resembling other arthropod venom peptides. The COATS pipeline identified six gene clusters under positive selection on branches leading to dune species, three of which reflected the preferred species tree. -

Final Project Completion Report
CEPF SMALL GRANT FINAL PROJECT COMPLETION REPORT Organization Legal Name: - Tarantula (Araneae: Theraphosidae) spider diversity, distribution and habitat-use: A study on Protected Area adequacy and Project Title: conservation planning at a landscape level in the Western Ghats of Uttara Kannada district, Karnataka Date of Report: 18 August 2011 Dr. Manju Siliwal Wildlife Information Liaison Development Society Report Author and Contact 9-A, Lal Bahadur Colony, Near Bharathi Colony Information Peelamedu Coimbatore 641004 Tamil Nadu, India CEPF Region: The Western Ghats Region (Sahyadri-Konkan and Malnad-Kodugu Corridors). 2. Strategic Direction: To improve the conservation of globally threatened species of the Western Ghats through systematic conservation planning and action. The present project aimed to improve the conservation status of two globally threatened (Molur et al. 2008b, Siliwal et al., 2008b) ground dwelling theraphosid species, Thrigmopoeus insignis and T. truculentus endemic to the Western Ghats through systematic conservation planning and action. Investment Priority 2.1 Monitor and assess the conservation status of globally threatened species with an emphasis on lesser-known organisms such as reptiles and fish. The present project was focused on an ignored or lesser-known group of spiders called Tarantulas/ Theraphosid spiders and provided valuable information on population status and potential conservation sites in Uttara Kannada district, which will help in future monitoring and assessment of conservation status of the two globally threatened theraphosid species T. insignis and Near Threatened T. truculentus. Investment Priority 2.3. Evaluate the existing protected area network for adequate globally threatened species representation and assess effectiveness of protected area types in biodiversity conservation. -

Tarantulas in the Pacific Northwest1
WSU Puyallup REC PLS-108 Updated July 2003 Tarantulas in the Pacific Northwest1 Tarantulas (Fig. 1) in the Pacific Northwest? Well, maybe not like the hairy monsters of the tropics, but some very interesting "atypical" species do occur here. Our species belong to the family Antrodiaetidae. One of our most common spiders is the folding-door spider, Antrodiaetus pacificus (Simon). It is a fairly large species, females ranging from 11 to 13 millimeters in length, males slightly smaller. They are generally dark brown to almost black in color with the abdomen purplish brown. Males are characterized by their long legs, slim bodies, and three tergites (hardened plates) on the abdomen. Females (Fig. 2) are more robust with only one tergite. These spiders excavate burrows in the soil or in damp, rotten wood, digging with a row of spines on each chelicer, known as a ratellum. The six to ten inch deep vertical shafts are lined with silk. The webbing extends beyond ground level as a short collar of camouflaged silk. The turret’s two sides may be drawn in by the occupant, forming two "doors" which meet in the middle. At night, Antrodiaetus assumes a foraging posture with its pedipalps and first pair of legs just touching the rim of silk at the mouth of the tube. In this position, the folding door spider can readily detect an insect moving above ground. The spider will leap out of its burrow with lightning speed, seize its victim, and drop back down, like a terrorizing Jack-in-the-box. When finished with its meal, it will add the insect's dry, dismembered body to a silk-covered trash pile at the bottom of its burrow. -

Distribution of Spiders in Coastal Grey Dunes
kaft_def 7/8/04 11:22 AM Pagina 1 SPATIAL PATTERNS AND EVOLUTIONARY D ISTRIBUTION OF SPIDERS IN COASTAL GREY DUNES Distribution of spiders in coastal grey dunes SPATIAL PATTERNS AND EVOLUTIONARY- ECOLOGICAL IMPORTANCE OF DISPERSAL - ECOLOGICAL IMPORTANCE OF DISPERSAL Dries Bonte Dispersal is crucial in structuring species distribution, population structure and species ranges at large geographical scales or within local patchily distributed populations. The knowledge of dispersal evolution, motivation, its effect on metapopulation dynamics and species distribution at multiple scales is poorly understood and many questions remain unsolved or require empirical verification. In this thesis we contribute to the knowledge of dispersal, by studying both ecological and evolutionary aspects of spider dispersal in fragmented grey dunes. Studies were performed at the individual, population and assemblage level and indicate that behavioural traits narrowly linked to dispersal, con- siderably show [adaptive] variation in function of habitat quality and geometry. Dispersal also determines spider distribution patterns and metapopulation dynamics. Consequently, our results stress the need to integrate knowledge on behavioural ecology within the study of ecological landscapes. / Promotor: Prof. Dr. Eckhart Kuijken [Ghent University & Institute of Nature Dries Bonte Conservation] Co-promotor: Prf. Dr. Jean-Pierre Maelfait [Ghent University & Institute of Nature Conservation] and Prof. Dr. Luc lens [Ghent University] Date of public defence: 6 February 2004 [Ghent University] Universiteit Gent Faculteit Wetenschappen Academiejaar 2003-2004 Distribution of spiders in coastal grey dunes: spatial patterns and evolutionary-ecological importance of dispersal Verspreiding van spinnen in grijze kustduinen: ruimtelijke patronen en evolutionair-ecologisch belang van dispersie door Dries Bonte Thesis submitted in fulfilment of the requirements for the degree of Doctor [Ph.D.] in Sciences Proefschrift voorgedragen tot het bekomen van de graad van Doctor in de Wetenschappen Promotor: Prof. -
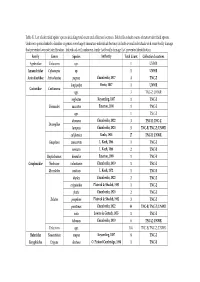
Table S1. List of Identified Spider Species Including Total Count and Collection Locations. Bold Cells Include Counts of Mature Identified Species
Table S1. List of identified spider species including total count and collection locations. Bold cells include counts of mature identified species. Unknown species linked to families or genera were largely immature individuals but may include several individuals with some bodily damage that prevented accurate identification. Individuals with unknown family had bodily damage that prevented identification. Family Genus Species Authority Total Count Collection Locations Agelenidae Unknown spp. 1 UNWR Amaurobiidae Cybaeopsis sp. 1 UNWR Antrodiaetidae Antrodiaetus pugnax Chamberlin, 1917 4 TNC-Z longipalpa Hentz, 1847 1 UNWR Corinnidae Castianeira spp. 3 TNC-Z; UNWR neglectus Keyserling, 1887 1 TNC-Z Drassodes saccatus Emerton, 1890 1 TNC-Z spp. 1 TNC-Z dromeus Chamberlin, 1922 3 TNC-B; TNC-Z Drassyllus lamprus Chamberlin, 1920 3 TNC-B; TNC-Z; UNWR californica Banks, 1904 17 TNC-B; UNWR Gnaphosa muscorum L. Koch, 1866 1 TNC-Z sericata L. Koch, 1866 2 TNC-B Haplodrassus hiemalis Emerton, 1909 1 TNC-B Gnaphosidae Nodocion voluntaries Chamberlin, 1919 1 TNC-Z Urozelotes rusticus L. Koch, 1872 1 TNC-B duplex Chamberlin, 1922 2 TNC-Z exiguioides Platnick & Shadab, 1983 1 TNC-Z fratis Chamberlin, 1920 2 TNC-Z Zelotes josephine Platnick & Shadab, 1983 3 TNC-Z puritanus Chamberlin, 1922 44 TNC-B; TNC-Z; UNWR sula Lowrie & Gertsch, 1955 1 TNC-Z tubuous Chamberlin, 1919 6 TNC-Z; UNWR Unknown spp. 184 TNC-B; TNC-Z; UNWR Hahniidae Neoantistea magna Keyserling, 1887 8 TNC-Z Linyphiidae Erigone dentosa O. Pickard-Cambridge, 1894 1 TNC-B spp. 1 TNC-Z Unknown spp. 13 TNC-Z; UNWR mccooki Montgomery, 1904 95 TNC-B; TNC-Z; UNWR Schizocosa minnesotensis Gertsch, 1934 25 TNC-B Lycosidae spp. -

A Preliminary Phylogeographic Study of the Diplurid Genus Microhexura
A PRELIMINARY PHYLOGEOGRAPHIC STUDY OF THE DIPLURID GENUS MICROHEXURA By Mia M. Martens A Thesis Submitted to the Faculty of the Graduate School of Western Carolina University in Partial Fulfillment of the Requirements for the Degree of Master of Science -,.L~:::::::.------~~r:=:::=""------ Dean of the Graduate School Fall 2005 Western Carolina University Cullowhee, North Carolina A PRELIMINARY PHYLOGEOGRAPHIC STUDY OF THE DIPLURID GENUS MICROHEXURA A thesis presented to the facuIty of the Graduate School of Western Carolina University in partial fulfillment of the requirements for the degree of Master of Science. By Mia M. Martens Director: Dr. James Costa H.P. and Katherine P. Robinson Professor of Biology Department of Biology December 2005 The Sequence A cross between a bug and a woolly worm Was here to seep away the sap. First, all the firs began to squirm, . Then fell to take a long nap. A change came to the forest, Darkness turned to light. The firs couldn't persist Due to this adelgid blight. Before too long, there was no shade. The light made the moss begin to dry, And as their populations began to fade, The spruce-fir moss spider started to sigh. Habitat loss and lack of lumber Caused questions for spiders to heed. What is our minimum number? How many of us do we need? Come; let's closely compare these three things. How's the weather, things like rain and snow? What's the rate of death to new beings? And finally, let's look at our gene flow. Now with this, we compare all our info. -

The Effects of Native and Non-Native Grasses on Spiders, Their Prey, and Their Interactions
Spiders in California’s grassland mosaic: The effects of native and non-native grasses on spiders, their prey, and their interactions by Kirsten Elise Hill A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Environmental Science, Policy, and Management in the GRADUATE DIVISION of the University of California, Berkeley Committee in charge: Professor Joe R. McBride, Chair Professor Rosemary G. Gillespie Professor Mary E. Power Spring 2014 © 2014 Abstract Spiders in California’s grassland mosaic: The effects of native and non-native grasses on spiders, their prey, and their interactions by Kirsten Elise Hill Doctor of Philosophy in Environmental Science and Policy Management University of California, Berkeley Professor Joe R. McBride, Chair Found in nearly all terrestrial ecosystems, small in size and able to occupy a variety of hunting niches, spiders’ consumptive effects on other arthropods can have important impacts for ecosystems. This dissertation describes research into spider populations and their interactions with potential arthropod prey in California’s native and non-native grasslands. In meadows found in northern California, native and non-native grassland patches support different functional groups of arthropod predators, sap-feeders, pollinators, and scavengers and arthropod diversity is linked to native plant diversity. Wandering spiders’ ability to forage within the meadow’s interior is linked to the distance from the shaded woodland boundary. Native grasses offer a cooler conduit into the meadow interior than non-native annual grasses during midsummer heat. Juvenile spiders in particular, are more abundant in the more structurally complex native dominated areas of the grassland. -

Tarantulas and Social Spiders
Tarantulas and Social Spiders: A Tale of Sex and Silk by Jonathan Bull BSc (Hons) MSc ICL Thesis Presented to the Institute of Biology of The University of Nottingham in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy The University of Nottingham May 2012 DEDICATION To my parents… …because they both said to dedicate it to the other… I dedicate it to both ii ACKNOWLEDGEMENTS First and foremost I would like to thank my supervisor Dr Sara Goodacre for her guidance and support. I am also hugely endebted to Dr Keith Spriggs who became my mentor in the field of RNA and without whom my understanding of the field would have been but a fraction of what it is now. Particular thanks go to Professor John Brookfield, an expert in the field of biological statistics and data retrieval. Likewise with Dr Susan Liddell for her proteomics assistance, a truly remarkable individual on par with Professor Brookfield in being able to simplify even the most complex techniques and analyses. Finally, I would really like to thank Janet Beccaloni for her time and resources at the Natural History Museum, London, permitting me access to the collections therein; ten years on and still a delight. Finally, amongst the greats, Alexander ‘Sasha’ Kondrashov… a true inspiration. I would also like to express my gratitude to those who, although may not have directly contributed, should not be forgotten due to their continued assistance and considerate nature: Dr Chris Wade (five straight hours of help was not uncommon!), Sue Buxton (direct to my bench creepy crawlies), Sheila Keeble (ventures and cleans where others dare not), Alice Young (read/checked my thesis and overcame her arachnophobia!) and all those in the Centre for Biomolecular Sciences.