Original Article Gene Expression Profiling Based on Microarray
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

The Role of Molecular Scaffolds at the Active Zone in Synaptic Vesicle Distribution and Release Probability
The Role of Molecular Scaffolds at the Active Zone in Synaptic Vesicle Distribution and Release Probability DISSERTATION To obtain the academic degree Doctor rerum naturalium (Dr. rer. nat.) submitted to the Department of Biology, Chemistry and Pharmacy Freie Universität Berlin Berlin, 2016 by Christina Beis (née Hollmann) This thesis was completed under the supervision of Prof. Dr. Stephan Sigrist from February 2011 to June 2016 at the Institute for Biology/ Genetics of Freie Universität Berlin, Germany. 1st reviewer: Prof. Dr. Stephan Sigrist 2nd reviewer: Prof. Dr. Hans-Joachim Pflüger Date of Defense: 21.11.2016 Statement of Authorship I hereby declare that the work presented in this thesis has been written independently and without inappropriate support. All sources of information are referenced. I hereby declare that this thesis has not been submitted, either in the same or in a different form, to this or any other university for a degree. ____________________________ Christina Beis Contents 1. Summary / Zusammenfassung .................................................................................................. 1 Summary....................................................................................................................................... 1 Zusammenfassung ........................................................................................................................ 3 2. Introduction .................................................................................................................................... -

Pharmacological Mechanisms Underlying the Hepatoprotective Effects of Ecliptae Herba on Hepatocellular Carcinoma
Hindawi Evidence-Based Complementary and Alternative Medicine Volume 2021, Article ID 5591402, 17 pages https://doi.org/10.1155/2021/5591402 Research Article Pharmacological Mechanisms Underlying the Hepatoprotective Effects of Ecliptae herba on Hepatocellular Carcinoma Botao Pan ,1 Wenxiu Pan ,2 Zheng Lu ,3 and Chenglai Xia 1,4 1Affiliated Foshan Maternity & Child Healthcare Hospital, Southern Medical University, Foshan 528000, China 2Department of Laboratory, Fifth People’s Hospital of Foshan, Foshan 528000, China 3Wuzhou Maternal and Child Health-Care Hospital, Wuzhou 543000, China 4School of Pharmaceutical Sciences, Southern Medical University, Guangzhou 510515, China Correspondence should be addressed to Zheng Lu; [email protected] and Chenglai Xia; [email protected] Received 30 January 2021; Revised 31 May 2021; Accepted 3 July 2021; Published 16 July 2021 Academic Editor: Hongcai Shang Copyright © 2021 Botao Pan et al. -is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. -e number of hepatocellular carcinoma (HCC) cases worldwide has increased significantly. As a traditional Chinese medicine (TCM) with a long history, Ecliptae herba (EH) has been widely used in HCC patients in China, but its hepatoprotective mechanism is still unclear. Methods. In this study, we applied a network pharmacology-based strategy and experimental ver- ification to systematically unravel the underlying mechanisms of EH against HCC. First, six active ingredients of EH were screened from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) by the ADME method. Subsequently, 52 potential targets of 6 active ingredients acting on HCC were screened from various databases, including TCMSP, DGIdb, SwissTargetPrediction, CTD, and GeneCards. -

Core Circadian Clock Transcription Factor BMAL1 Regulates Mammary Epithelial Cell
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.23.432439; this version posted February 23, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Title: Core circadian clock transcription factor BMAL1 regulates mammary epithelial cell 2 growth, differentiation, and milk component synthesis. 3 Authors: Theresa Casey1ǂ, Aridany Suarez-Trujillo1, Shelby Cummings1, Katelyn Huff1, 4 Jennifer Crodian1, Ketaki Bhide2, Clare Aduwari1, Kelsey Teeple1, Avi Shamay3, Sameer J. 5 Mabjeesh4, Phillip San Miguel5, Jyothi Thimmapuram2, and Karen Plaut1 6 Affiliations: 1. Department of Animal Science, Purdue University, West Lafayette, IN, USA; 2. 7 Bioinformatics Core, Purdue University; 3. Animal Science Institute, Agriculture Research 8 Origination, The Volcani Center, Rishon Letsiyon, Israel. 4. Department of Animal Sciences, 9 The Robert H. Smith Faculty of Agriculture, Food, and Environment, The Hebrew University of 10 Jerusalem, Rehovot, Israel. 5. Genomics Core, Purdue University 11 Grant support: Binational Agricultural Research Development (BARD) Research Project US- 12 4715-14; Photoperiod effects on milk production in goats: Are they mediated by the molecular 13 clock in the mammary gland? 14 ǂAddress for correspondence. 15 Theresa M. Casey 16 BCHM Room 326 17 175 South University St. 18 West Lafayette, IN 47907 19 Email: [email protected] 20 Phone: 802-373-1319 21 22 bioRxiv preprint doi: https://doi.org/10.1101/2021.02.23.432439; this version posted February 23, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Ion Channels 3 1
r r r Cell Signalling Biology Michael J. Berridge Module 3 Ion Channels 3 1 Module 3 Ion Channels Synopsis Ion channels have two main signalling functions: either they can generate second messengers or they can function as effectors by responding to such messengers. Their role in signal generation is mainly centred on the Ca2 + signalling pathway, which has a large number of Ca2+ entry channels and internal Ca2+ release channels, both of which contribute to the generation of Ca2 + signals. Ion channels are also important effectors in that they mediate the action of different intracellular signalling pathways. There are a large number of K+ channels and many of these function in different + aspects of cell signalling. The voltage-dependent K (KV) channels regulate membrane potential and + excitability. The inward rectifier K (Kir) channel family has a number of important groups of channels + + such as the G protein-gated inward rectifier K (GIRK) channels and the ATP-sensitive K (KATP) + + channels. The two-pore domain K (K2P) channels are responsible for the large background K current. Some of the actions of Ca2 + are carried out by Ca2+-sensitive K+ channels and Ca2+-sensitive Cl − channels. The latter are members of a large group of chloride channels and transporters with multiple functions. There is a large family of ATP-binding cassette (ABC) transporters some of which have a signalling role in that they extrude signalling components from the cell. One of the ABC transporters is the cystic − − fibrosis transmembrane conductance regulator (CFTR) that conducts anions (Cl and HCO3 )and contributes to the osmotic gradient for the parallel flow of water in various transporting epithelia. -

Gene Expression Profiles of Patients with Cerebral Hematoma Following Spontaneous Intracerebral Hemorrhage
MOLECULAR MEDICINE REPORTS 10: 1671-1678, 2014 Gene expression profiles of patients with cerebral hematoma following spontaneous intracerebral hemorrhage TAO YANG1,2, JIANWEN GU1, BIN KONG1,2, YONGQIN KUANG1, LIN CHENG1, JINGMIN CHENG1,2, XUN XIA1,2, YUAN MA1,2 and JUNHAI ZHANG1 1Department of Neurosurgery, Chengdu Military General Hospital, Chengdu, Sichuan 610083; 2Department of Postgraduate, Third Military Medical University, Chongqing 400038, P.R. China Received October 13, 2013; Accepted March 27, 2014 DOI: 10.3892/mmr.2014.2421 Abstract. The present study aimed to investigate the gene ITPR1-RYR2, CAMK2A-RYR2 and ITGA5-LAMB1 interac- functions and expression profiles in perihematomal (PH) tion pairs. The present study provides several potential targets brain regions following spontaneous intracerebral hemor- to decrease hematoma expansion and alleviate neuronal cell rhage. The gene expression profiles were downloaded from death following spontaneous intracerebral hemorrhage. the Gene Expression Omnibus database under accession number GSE24265, which includes 11 brain samples from Introduction different regions, including four samples from PH areas, four from contralateral grey matter (CG) and three from contra- Spontaneous intracerebral hemorrhage (ICH) involves the lateral white matter (CW). The gene expression profiles were extravasation of blood within the brain parenchyma in the pre-processed and the differentially expressed genes (DEGs) absence of trauma or surgery, accounting for 10-30% of all between PH and CG tissue, and PH and CW tissue were identi- strokes worldwide (1). ICH is a complex, dynamic process fied using R packages. The expression of genes in different that consists of three distinct phases (2): i) Initial hemorrhage, tissues was analyzed by hierarchical clustering. -
Primepcr™Assay Validation Report
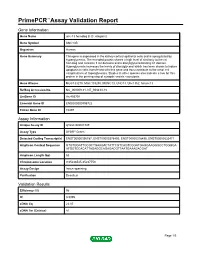
PrimePCR™Assay Validation Report Gene Information Gene Name unc-13 homolog B (C. elegans) Gene Symbol UNC13B Organism Human Gene Summary This gene is expressed in the kidney cortical epithelial cells and is upregulated by hyperglycemia. The encoded protein shares a high level of similarity to the rat homolog and contains 3 C2 domains and a diacylglycerol-binding C1 domain. Hyperglycemia increases the levels of diacylglycerol which has been shown to induce apoptosis in cells transfected with this gene and thus contribute to the renal cell complications of hyperglycemia. Studies in other species also indicate a role for this protein in the priming step of synaptic vesicle exocytosis. Gene Aliases MGC133279, MGC133280, MUNC13, UNC13, Unc13h2, hmunc13 RefSeq Accession No. NC_000009.11, NT_008413.18 UniGene ID Hs.493791 Ensembl Gene ID ENSG00000198722 Entrez Gene ID 10497 Assay Information Unique Assay ID qHsaCID0007349 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000396787, ENST00000378495, ENST00000378496, ENST00000535471 Amplicon Context Sequence GTGTGGATTGCGCTGAAGACTATTCGTCAGTCGGATGAGGAAGGGCCTGGGGA ATGGTCCACATTAGAGGCAGAGACGTTAATGAAAGACGAT Amplicon Length (bp) 63 Chromosome Location 9:35236545-35237753 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 96 R2 0.9995 cDNA Cq 22.07 cDNA Tm (Celsius) 81 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report -

I REGENERATIVE MEDICINE APPROACHES to SPINAL CORD
REGENERATIVE MEDICINE APPROACHES TO SPINAL CORD INJURY A Dissertation Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Ashley Elizabeth Mohrman March 2017 i ABSTRACT Hundreds of thousands of people suffer from spinal cord injuries in the U.S.A. alone, with very few patients ever experiencing complete recovery. Complexity of the tissue and inflammatory response contribute to this lack of recovery, as the proper function of the central nervous system relies on its highly specific structural and spatial organization. The overall goal of this dissertation project is to study the central nervous system in the healthy and injured state so as to devise appropriate strategies to recover tissue homeostasis, and ultimately function, from an injured state. A specific spinal cord injury model, syringomyelia, was studied; this condition presents as a fluid filled cyst within the spinal cord. Molecular evaluation at three and six weeks post-injury revealed a large inflammatory response including leukocyte invasion, losses in neuronal transmission and signaling, and upregulation in important osmoregulators. These included osmotic stress regulating metabolites betaine and taurine, as well as the betaine/GABA transporter (BGT-1), potassium chloride transporter (KCC4), and water transporter aquaporin 1 (AQP1). To study cellular behavior in native tissue, adult neural stem cells from the subventricular niche were differentiated in vitro. These cells were tested under various culture conditions for cell phenotype preferences. A mostly pure (>80%) population of neural stem cells could be specified using soft, hydrogel substrates with a laminin coating and interferon-γ supplementation. -

Role of Kcnma1 in the Pathogenesis of Gepd Syndrome
ROLE OF KCNMA1 IN THE PATHOGENESIS OF GEPD SYNDROME WEI DU Bachelor of Science in Biochemistry Beijing Normal University June, 2001 Submitted in partial fulfillment of requirements for the degree DOCTOR OF PHILOSOPHY IN REGULATORY BIOLOGY at the CLEVELAND STATE UNIVERSITY NOVEMBER, 2009 This dissertation has been approved for the Department of Biological, Geological, and Environmental Sciences and the College of Graduate Studies by ___________________________________________ Dissertation Chairperson, Qing Wang __________________________ Department & Date ___________________________________________ Crystal Weyman __________________________ Department & Date ___________________________________________ Sadashiva Karnik __________________________ Department & Date ___________________________________________ Xiaoxia Li __________________________ Department & Date ___________________________________________ Roman Kondratov __________________________ Department & Date ___________________________________________ Qiuyun Chen __________________________ Department & Date ACKNOWLEDGEMENT I would like to thank my advisor, Dr. Qing Wang, for his help and support. I want to thank my committee members, Dr. Crystal Weyman, Dr. Sadashiva Karnik, Dr. Xiaoxia Li, Dr. Roman Kondratov, and Dr. Qiuyun Chen, for their time and effort. I want to thank Dr. Jocelyn Bautista for collecting the blood samples from the patients. I want to thank our collaborators, Dr. Jianmin Cui, Dr. George Richerson, Dr. Huanghe Yang, and Dr. Ana Diez-Sampedro. I want to thank Dr. -

Reduced Insulin Secretion Correlates with Decreased Expression of Exocytotic Genes in Pancreatic Islets from Patients with Type 2 Diabetes
Molecular and Cellular Endocrinology 364 (2012) 36–45 Contents lists available at SciVerse ScienceDirect Molecular and Cellular Endocrinology journal homepage: www.elsevier.com/locate/mce Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes Sofia A. Andersson a, Anders H. Olsson b, Jonathan L.S. Esguerra a, Emilia Heimann e, Claes Ladenvall c, Anna Edlund a, Albert Salehi d, Jalal Taneera c, Eva Degerman e, Leif Groop c, Charlotte Ling b, ⇑ Lena Eliasson a, a Islet Cell Exocytosis, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden b Epigenetics and Diabetes, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden c Diabetes and Endocrinology, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden d Islet Cell Physiology, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden e Department of Experimental Medical Sciences, Biomedical Center, Lund University, Lund, Sweden article info abstract Article history: Reduced insulin release has been linked to defect exocytosis in b-cells. However, whether expression of Received 14 December 2011 genes suggested to be involved in the exocytotic process (exocytotic genes) is altered in pancreatic islets Received in revised form 7 August 2012 from patients with type 2 diabetes (T2D), and correlate to insulin secretion, needs to be further investi- Accepted 13 August 2012 gated. Available online 23 August 2012 Analysing expression levels of 23 exocytotic genes using microarray revealed reduced expression of five genes in human T2D islets (v2 = 13.25; p < 0.001). -

Genomic and Transcriptome Analysis Revealing an Oncogenic Functional Module in Meningiomas
Neurosurg Focus 35 (6):E3, 2013 ©AANS, 2013 Genomic and transcriptome analysis revealing an oncogenic functional module in meningiomas XIAO CHANG, PH.D.,1 LINGLING SHI, PH.D.,2 FAN GAO, PH.D.,1 JONATHAN RUssIN, M.D.,3 LIYUN ZENG, PH.D.,1 SHUHAN HE, B.S.,3 THOMAS C. CHEN, M.D.,3 STEVEN L. GIANNOTTA, M.D.,3 DANIEL J. WEISENBERGER, PH.D.,4 GAbrIEL ZADA, M.D.,3 KAI WANG, PH.D.,1,5,6 AND WIllIAM J. MAck, M.D.1,3 1Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, Los Angeles, California; 2GHM Institute of CNS Regeneration, Jinan University, Guangzhou, China; 3Department of Neurosurgery, Keck School of Medicine, University of Southern California, Los Angeles, California; 4USC Epigenome Center, Keck School of Medicine, University of Southern California, Los Angeles, California; 5Department of Psychiatry, Keck School of Medicine, University of Southern California, Los Angeles, California; and 6Division of Bioinformatics, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California Object. Meningiomas are among the most common primary adult brain tumors. Although typically benign, roughly 2%–5% display malignant pathological features. The key molecular pathways involved in malignant trans- formation remain to be determined. Methods. Illumina expression microarrays were used to assess gene expression levels, and Illumina single- nucleotide polymorphism arrays were used to identify copy number variants in benign, atypical, and malignant me- ningiomas (19 tumors, including 4 malignant ones). The authors also reanalyzed 2 expression data sets generated on Affymetrix microarrays (n = 68, including 6 malignant ones; n = 56, including 3 malignant ones). -

Whole-Genome Sequencing Reveals Genetic Variation in the Asian House Rat
INVESTIGATION Whole-Genome Sequencing Reveals Genetic Variation in the Asian House Rat Huajing Teng,*,†,‡ Yaohua Zhang,* Chengmin Shi,*,§ Fengbiao Mao,‡ Lingling Hou,‡ Hongling Guo,*,† Zhongsheng Sun,‡ and Jianxu Zhang*,1 *The State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, 100101 Beijing, China, †University of Chinese Academy of Sciences, 100049 Beijing, China, § ‡ Beijing Institutes of Life Science, Chinese Academy of Sciences, 100101 Beijing, China, and Beijing Institute of Genomics, Chinese Academy of Sciences, 100101 Beijing, China ABSTRACT Whole-genome sequencing of wild-derived rat species can provide novel genomic resources, KEYWORDS which may help decipher the genetics underlying complex phenotypes. As a notorious pest, reservoir of human genetic pathogens, and colonizer, the Asian house rat, Rattus tanezumi, is successfully adapted to its habitat. However, landscape little is known regarding genetic variation in this species. In this study, we identified over 41,000,000 single- next-generation nucleotide polymorphisms, plus insertions and deletions, through whole-genome sequencing and bioinfor- sequencing matics analyses. Moreover, we identified over 12,000 structural variants, including 143 chromosomal inversions. Rattus tanezumi Further functional analyses revealed several fixed nonsense mutations associated with infection and immunity- single-nucleotide related adaptations, and a number of fixed missense mutations that may be related to anticoagulant resistance. polymorphisms A genome-wide scan for loci under selection identified various genes related to neural activity. Our whole- structural genome sequencing data provide a genomic resource for future genetic studies of the Asian house rat species variations and have the potential to facilitate understanding of the molecular adaptations of rats to their ecological niches.