Download Download
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of Molecular Scaffolds at the Active Zone in Synaptic Vesicle Distribution and Release Probability
The Role of Molecular Scaffolds at the Active Zone in Synaptic Vesicle Distribution and Release Probability DISSERTATION To obtain the academic degree Doctor rerum naturalium (Dr. rer. nat.) submitted to the Department of Biology, Chemistry and Pharmacy Freie Universität Berlin Berlin, 2016 by Christina Beis (née Hollmann) This thesis was completed under the supervision of Prof. Dr. Stephan Sigrist from February 2011 to June 2016 at the Institute for Biology/ Genetics of Freie Universität Berlin, Germany. 1st reviewer: Prof. Dr. Stephan Sigrist 2nd reviewer: Prof. Dr. Hans-Joachim Pflüger Date of Defense: 21.11.2016 Statement of Authorship I hereby declare that the work presented in this thesis has been written independently and without inappropriate support. All sources of information are referenced. I hereby declare that this thesis has not been submitted, either in the same or in a different form, to this or any other university for a degree. ____________________________ Christina Beis Contents 1. Summary / Zusammenfassung .................................................................................................. 1 Summary....................................................................................................................................... 1 Zusammenfassung ........................................................................................................................ 3 2. Introduction .................................................................................................................................... -

Pharmacological Mechanisms Underlying the Hepatoprotective Effects of Ecliptae Herba on Hepatocellular Carcinoma
Hindawi Evidence-Based Complementary and Alternative Medicine Volume 2021, Article ID 5591402, 17 pages https://doi.org/10.1155/2021/5591402 Research Article Pharmacological Mechanisms Underlying the Hepatoprotective Effects of Ecliptae herba on Hepatocellular Carcinoma Botao Pan ,1 Wenxiu Pan ,2 Zheng Lu ,3 and Chenglai Xia 1,4 1Affiliated Foshan Maternity & Child Healthcare Hospital, Southern Medical University, Foshan 528000, China 2Department of Laboratory, Fifth People’s Hospital of Foshan, Foshan 528000, China 3Wuzhou Maternal and Child Health-Care Hospital, Wuzhou 543000, China 4School of Pharmaceutical Sciences, Southern Medical University, Guangzhou 510515, China Correspondence should be addressed to Zheng Lu; [email protected] and Chenglai Xia; [email protected] Received 30 January 2021; Revised 31 May 2021; Accepted 3 July 2021; Published 16 July 2021 Academic Editor: Hongcai Shang Copyright © 2021 Botao Pan et al. -is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. -e number of hepatocellular carcinoma (HCC) cases worldwide has increased significantly. As a traditional Chinese medicine (TCM) with a long history, Ecliptae herba (EH) has been widely used in HCC patients in China, but its hepatoprotective mechanism is still unclear. Methods. In this study, we applied a network pharmacology-based strategy and experimental ver- ification to systematically unravel the underlying mechanisms of EH against HCC. First, six active ingredients of EH were screened from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) by the ADME method. Subsequently, 52 potential targets of 6 active ingredients acting on HCC were screened from various databases, including TCMSP, DGIdb, SwissTargetPrediction, CTD, and GeneCards. -

The Pathogenetic Role of Β-Cell Mitochondria in Type 2 Diabetes
236 3 Journal of M Fex et al. Mitochondria in β-cells 236:3 R145–R159 Endocrinology REVIEW The pathogenetic role of β-cell mitochondria in type 2 diabetes Malin Fex1, Lisa M Nicholas1, Neelanjan Vishnu1, Anya Medina1, Vladimir V Sharoyko1, David G Nicholls1, Peter Spégel1,2 and Hindrik Mulder1 1Department of Clinical Sciences in Malmö, Unit of Molecular Metabolism, Lund University Diabetes Centre, Clinical Research Center, Malmö University Hospital, Lund University, Malmö, Sweden 2Department of Chemistry, Center for Analysis and Synthesis, Lund University, Sweden Correspondence should be addressed to H Mulder: [email protected] Abstract Mitochondrial metabolism is a major determinant of insulin secretion from pancreatic Key Words β-cells. Type 2 diabetes evolves when β-cells fail to release appropriate amounts f TCA cycle of insulin in response to glucose. This results in hyperglycemia and metabolic f coupling signal dysregulation. Evidence has recently been mounting that mitochondrial dysfunction f oxidative phosphorylation plays an important role in these processes. Monogenic dysfunction of mitochondria is a f mitochondrial transcription rare condition but causes a type 2 diabetes-like syndrome owing to β-cell failure. Here, f genetic variation we describe novel advances in research on mitochondrial dysfunction in the β-cell in type 2 diabetes, with a focus on human studies. Relevant studies in animal and cell models of the disease are described. Transcriptional and translational regulation in mitochondria are particularly emphasized. The role of metabolic enzymes and pathways and their impact on β-cell function in type 2 diabetes pathophysiology are discussed. The role of genetic variation in mitochondrial function leading to type 2 diabetes is highlighted. -

SARS-Cov-2 Restructures the Host Chromatin Architecture 2
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.20.453146; this version posted July 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 SARS-CoV-2 Restructures the Host Chromatin Architecture 2 3 Authors: Ruoyu Wang 1,2,†, Joo-Hyung Lee 1,†, Feng Xiong 1, Jieun Kim 3,4, Lana Al Hasani 1,2, Xiaoyi 4 Yuan 3,4, Pooja Shivshankar 1,3,4, Joanna Krakowiak 1, Chuangye Qi 1, Yanyu Wang 3,4, Holger K. 5 Eltzschig 2,3,4, Wenbo Li 1,2,* 6 7 Affiliations: 8 1 Department of Biochemistry and Molecular Biology, McGovern Medical School, University of Texas 9 Health Science Center, Houston, 77030, TX, USA 10 2 Graduate School of Biomedical Sciences, University of Texas MD Anderson Cancer Center and 11 UTHealth, Houston, 77030, TX, USA 12 3 Department of Anesthesiology, McGovern Medical School, University of Texas Health Science Center, 13 Houston, 77030, TX, USA 14 4 Center for Perioperative Medicine, McGovern Medical School, University of Texas Health Science 15 Center, Houston, 77030, TX, USA 16 17 † These authors contributed equally 18 * Correspondence: Wenbo Li, Ph.D. ([email protected]). MSB 6.161, Fannin Street, Houston, 19 Texas, 77030, USA; Tel: 713-500-6103. 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.20.453146; this version posted July 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

NDUFB6 (NM 182739) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC219791 NDUFB6 (NM_182739) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: NDUFB6 (NM_182739) Human Tagged ORF Clone Tag: Myc-DDK Symbol: NDUFB6 Synonyms: B17; CI Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC219791 representing NM_182739 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGACGGGGTACACTCCGGATGAGAAACTGCGGCTGCAGCAGCTGCGAGAGCTGAGAAGGCGATGGCTGA AGGACCAGGAGCTGAGCCCTCGGGAGCCGGTGCTGCCCCCACAGAAGATGGGGCCTATGGAGAAATTCTG GAATAAATTTTTGGAGAATAAATCCCCTTGGAGGAAAATGGTCCATGGGGTATACAAAAAGAGTATCTTT GTTTTCACTCATGTACTTGTACCTGTCTGGATTATTCATTATTACATGAAGTATCATGTTTCTGGTGATA CAATTCTGGAGACTGGAGAAGTAATTCCACCAATGAAAGAATTTCCTGATCAACATCAT ACGCGTACGCGGCCGCTCGAGCAGAAACTCATCTCAGAAGAGGATCTGGCAGCAAATGATATCCTGGATT ACAAGGATGACGACGATAAGGTTTAA Protein Sequence: >RC219791 representing NM_182739 Red=Cloning site Green=Tags(s) MTGYTPDEKLRLQQLRELRRRWLKDQELSPREPVLPPQKMGPMEKFWNKFLENKSPWRKMVHGVYKKSIF VFTHVLVPVWIIHYYMKYHVSGDTILETGEVIPPMKEFPDQHH TRTRPLEQKLISEEDLAANDILDYKDDDDKV Restriction Sites: SgfI-MluI This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, -
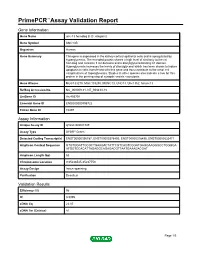
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name unc-13 homolog B (C. elegans) Gene Symbol UNC13B Organism Human Gene Summary This gene is expressed in the kidney cortical epithelial cells and is upregulated by hyperglycemia. The encoded protein shares a high level of similarity to the rat homolog and contains 3 C2 domains and a diacylglycerol-binding C1 domain. Hyperglycemia increases the levels of diacylglycerol which has been shown to induce apoptosis in cells transfected with this gene and thus contribute to the renal cell complications of hyperglycemia. Studies in other species also indicate a role for this protein in the priming step of synaptic vesicle exocytosis. Gene Aliases MGC133279, MGC133280, MUNC13, UNC13, Unc13h2, hmunc13 RefSeq Accession No. NC_000009.11, NT_008413.18 UniGene ID Hs.493791 Ensembl Gene ID ENSG00000198722 Entrez Gene ID 10497 Assay Information Unique Assay ID qHsaCID0007349 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000396787, ENST00000378495, ENST00000378496, ENST00000535471 Amplicon Context Sequence GTGTGGATTGCGCTGAAGACTATTCGTCAGTCGGATGAGGAAGGGCCTGGGGA ATGGTCCACATTAGAGGCAGAGACGTTAATGAAAGACGAT Amplicon Length (bp) 63 Chromosome Location 9:35236545-35237753 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 96 R2 0.9995 cDNA Cq 22.07 cDNA Tm (Celsius) 81 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report -

THE FUNCTIONAL SIGNIFICANCE of MITOCHONDRIAL SUPERCOMPLEXES in C. ELEGANS by WICHIT SUTHAMMARAK Submitted in Partial Fulfillment
THE FUNCTIONAL SIGNIFICANCE OF MITOCHONDRIAL SUPERCOMPLEXES in C. ELEGANS by WICHIT SUTHAMMARAK Submitted in partial fulfillment of the requirements For the degree of Doctor of Philosophy Dissertation Advisor: Drs. Margaret M. Sedensky & Philip G. Morgan Department of Genetics CASE WESTERN RESERVE UNIVERSITY January, 2011 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of _____________________________________________________ candidate for the ______________________degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. Dedicated to my family, my teachers and all of my beloved ones for their love and support ii ACKNOWLEDGEMENTS My advanced academic journey began 5 years ago on the opposite side of the world. I traveled to the United States from Thailand in search of a better understanding of science so that one day I can return to my homeland and apply the knowledge and experience I have gained to improve the lives of those affected by sickness and disease yet unanswered by science. Ultimately, I hoped to make the academic transition into the scholarly community by proving myself through scientific research and understanding so that I can make a meaningful contribution to both the scientific and medical communities. The following dissertation would not have been possible without the help, support, and guidance of a lot of people both near and far. I wish to thank all who have aided me in one way or another on this long yet rewarding journey. My sincerest thanks and appreciation goes to my advisors Philip Morgan and Margaret Sedensky. -

Reduced Insulin Secretion Correlates with Decreased Expression of Exocytotic Genes in Pancreatic Islets from Patients with Type 2 Diabetes
Molecular and Cellular Endocrinology 364 (2012) 36–45 Contents lists available at SciVerse ScienceDirect Molecular and Cellular Endocrinology journal homepage: www.elsevier.com/locate/mce Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes Sofia A. Andersson a, Anders H. Olsson b, Jonathan L.S. Esguerra a, Emilia Heimann e, Claes Ladenvall c, Anna Edlund a, Albert Salehi d, Jalal Taneera c, Eva Degerman e, Leif Groop c, Charlotte Ling b, ⇑ Lena Eliasson a, a Islet Cell Exocytosis, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden b Epigenetics and Diabetes, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden c Diabetes and Endocrinology, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden d Islet Cell Physiology, Lund University Diabetes Centre, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden e Department of Experimental Medical Sciences, Biomedical Center, Lund University, Lund, Sweden article info abstract Article history: Reduced insulin release has been linked to defect exocytosis in b-cells. However, whether expression of Received 14 December 2011 genes suggested to be involved in the exocytotic process (exocytotic genes) is altered in pancreatic islets Received in revised form 7 August 2012 from patients with type 2 diabetes (T2D), and correlate to insulin secretion, needs to be further investi- Accepted 13 August 2012 gated. Available online 23 August 2012 Analysing expression levels of 23 exocytotic genes using microarray revealed reduced expression of five genes in human T2D islets (v2 = 13.25; p < 0.001). -

Short-Term Exercise Training Does Not Stimulate Skeletal Muscle ATP Synthesis in Relatives of Humans with Type 2 Diabetes
Diabetes Publish Ahead of Print, published online March 5, 2009 Short-term exercise training does not stimulate skeletal muscle ATP synthesis in relatives of humans with type 2 diabetes Gertrud Kacerovsky-Bielesz1,2, Marek Chmelik2,3, Charlotte Ling4, Rochus Pokan5, Julia Szendroedi1,2, Michaela Farukuoye2, Michaela Kacerovsky2, Albrecht I. Schmid2,3, Stephan Gruber3, Michael Wolzt6, Ewald Moser3, Giovanni Pacini7, Gerhard Smekal5, Leif Groop4, Michael Roden1,2,8 1 1. Medical Department, Hanusch Hospital, Vienna, Austria, 2 Karl-Landsteiner Institute for Endocrinology and Metabolism, Vienna, Austria, 3 MR Center of Excellence, Medical University of Vienna, Austria, 4 Department of Clinical Sciences, Lund University, Malmö, Sweden, 5 Department of Sports and Exercise Physiology, University of Vienna, Austria, 6 Department of Clinical Pharmacology, Medical University of Vienna, Austria, 7 Metabolic Unit, Institute of Biomedical Engineering, CNR, Padova, Italy 8 Institute for Clinical Diabetology, German Diabetes Center - Leibniz Center for Diabetes Research, Department of Medicine/Metabolic Diseases, Heinrich Heine University Düsseldorf, Düsseldorf, Germany Corresponding Author: Michael Roden, MD Email: [email protected] Clinical trial reg. No. NCT00710008, clinicaltrials.gov. Additional information for this article can be found in an online appendix at http://diabetes.diabetesjournals.org Submitted 6 September 2008 and accepted 27 February 2009. This is an uncopyedited electronic version of an article accepted for publication in Diabetes. The American Diabetes Association, publisher of Diabetes, is not responsible for any errors or omissions in this version of the manuscript or any version derived from it by third parties. The definitive publisher-authenticated version will be available in a future issue of Diabetes in print and online at http://diabetes.diabetesjournals.org. -

Genomic and Transcriptome Analysis Revealing an Oncogenic Functional Module in Meningiomas
Neurosurg Focus 35 (6):E3, 2013 ©AANS, 2013 Genomic and transcriptome analysis revealing an oncogenic functional module in meningiomas XIAO CHANG, PH.D.,1 LINGLING SHI, PH.D.,2 FAN GAO, PH.D.,1 JONATHAN RUssIN, M.D.,3 LIYUN ZENG, PH.D.,1 SHUHAN HE, B.S.,3 THOMAS C. CHEN, M.D.,3 STEVEN L. GIANNOTTA, M.D.,3 DANIEL J. WEISENBERGER, PH.D.,4 GAbrIEL ZADA, M.D.,3 KAI WANG, PH.D.,1,5,6 AND WIllIAM J. MAck, M.D.1,3 1Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, Los Angeles, California; 2GHM Institute of CNS Regeneration, Jinan University, Guangzhou, China; 3Department of Neurosurgery, Keck School of Medicine, University of Southern California, Los Angeles, California; 4USC Epigenome Center, Keck School of Medicine, University of Southern California, Los Angeles, California; 5Department of Psychiatry, Keck School of Medicine, University of Southern California, Los Angeles, California; and 6Division of Bioinformatics, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California Object. Meningiomas are among the most common primary adult brain tumors. Although typically benign, roughly 2%–5% display malignant pathological features. The key molecular pathways involved in malignant trans- formation remain to be determined. Methods. Illumina expression microarrays were used to assess gene expression levels, and Illumina single- nucleotide polymorphism arrays were used to identify copy number variants in benign, atypical, and malignant me- ningiomas (19 tumors, including 4 malignant ones). The authors also reanalyzed 2 expression data sets generated on Affymetrix microarrays (n = 68, including 6 malignant ones; n = 56, including 3 malignant ones). -

Whole-Genome Sequencing Reveals Genetic Variation in the Asian House Rat
INVESTIGATION Whole-Genome Sequencing Reveals Genetic Variation in the Asian House Rat Huajing Teng,*,†,‡ Yaohua Zhang,* Chengmin Shi,*,§ Fengbiao Mao,‡ Lingling Hou,‡ Hongling Guo,*,† Zhongsheng Sun,‡ and Jianxu Zhang*,1 *The State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, 100101 Beijing, China, †University of Chinese Academy of Sciences, 100049 Beijing, China, § ‡ Beijing Institutes of Life Science, Chinese Academy of Sciences, 100101 Beijing, China, and Beijing Institute of Genomics, Chinese Academy of Sciences, 100101 Beijing, China ABSTRACT Whole-genome sequencing of wild-derived rat species can provide novel genomic resources, KEYWORDS which may help decipher the genetics underlying complex phenotypes. As a notorious pest, reservoir of human genetic pathogens, and colonizer, the Asian house rat, Rattus tanezumi, is successfully adapted to its habitat. However, landscape little is known regarding genetic variation in this species. In this study, we identified over 41,000,000 single- next-generation nucleotide polymorphisms, plus insertions and deletions, through whole-genome sequencing and bioinfor- sequencing matics analyses. Moreover, we identified over 12,000 structural variants, including 143 chromosomal inversions. Rattus tanezumi Further functional analyses revealed several fixed nonsense mutations associated with infection and immunity- single-nucleotide related adaptations, and a number of fixed missense mutations that may be related to anticoagulant resistance. polymorphisms A genome-wide scan for loci under selection identified various genes related to neural activity. Our whole- structural genome sequencing data provide a genomic resource for future genetic studies of the Asian house rat species variations and have the potential to facilitate understanding of the molecular adaptations of rats to their ecological niches. -

The Association of UNC13B Gene Polymorphisms and Diabetic
CLINICAL RESEARCH e-ISSN 1643-3750 © Med Sci Monit, 2019; 25: 8527-8533 DOI: 10.12659/MSM.919930 Received: 2019.09.05 Accepted: 2019.10.21 The Association of UNC13B Gene Polymorphisms Published: 2019.11.12 and Diabetic Kidney Disease in a Chinese Han Population Authors’ Contribution: AEG 1 Ya Wang 1 Department of Endocrinology, Jingzhou First People’s Hospital, The First Affiliated Study Design A AC 2 Jie Tan Hospital of Yangtze University, Jingzhou, Hubei, P.R. China Data Collection B 2 Department of Hematology, Jingzhou First People’s Hospital, The First Affiliated Statistical Analysis C BF 1 Dan Liu Hospital of Yangtze University, Jingzhou, Hubei, P.R .China Data Interpretation D BD 3 Yameng Yang 3 Department of Rheumatism and Immunology, Jingzhou First People’s Hospital, Manuscript Preparation E CG 1 Hongyan Wu The First Affiliated Hospital of Yangtze University, Jingzhou, Hubei, P.R. China Literature Search F Funds Collection G Corresponding Authors: Jie Tan, e-mail: [email protected], Hongyan Wu, e-mail: [email protected] Source of support: This study was supported by the Science and Technology Planning Project of Jingzhou Science and Technology Bureau, China (Grant No. 2018073) Background: Polymorphisms in the UNC13B gene are associated with diabetic kidney disease (DKD) in the European pop- ulation. Asian populations are more likely to suffer from complications of type 2 diabetes mellitus (T2DM), including diabetic kidney disease (DKD). This case-control study aimed to investigate the association between UNC13B gene polymorphisms and DKD in a Chinese Han population. Material/Methods: Five single nucleotide polymorphism (SNP) loci (rs13293564, rs17360668, rs10114937, rs661712, and rs2281999) were genotyped in the UNC13B gene in 600 Chinese Han subjects.