Amaia Caro Aramendia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Spatial Predictive Distribution Modelling of Madeira's Endemic
DEPARTAMENTO DE ZOOLOGIA FACULDADE DE CIÊNCIAS E TECNOLOGIA UNIVERSIDADE DE COIMBRA Spatial predictive distribution modelling of Madeira’s endemic land snail species Dinarte Nuno Freitas Teixeira 2009 REGIÃO AUTÓNOMA DA REPÚBLICA PORTUGUESA UNIÃO EUROPEIA MADEIRA FSE DEPARTAMENTO DE ZOOLOGIA FACULDADE DE CIÊNCIAS E TECNOLOGIA UNIVERSIDADE DE COIMBRA Spatial predictive distribution modelling of Madeira’s endemic land snail species Dissertação apresentada à Universidade de Coimbra para cumprimento dos requisitos necessários à obtenção do grau de Mestre em Ecologia, realizada sob a orientação científica do Professor Doutor José Paulo Sousa (Universidade de Coimbra) e do Professor Doutor José Manuel Jesus (Universidade da Madeira). Dinarte Nuno Freitas Teixeir a 2009 O presente trabalho foi financiado pelo Centro de Ciência e Tecnologia da Madeira (CITMA), através da bolsa de Mestrado FSE BM I/2008 – 531, ao abrigo do Programa Operacional de Valorização do Potencial Humano e Coesão Social da RAM (RUMOS). REGIÃO AUTÓNOMA DA MADEIRA REPÚBLICA PORTUGUESA UNIÃO EUROPEIA FSE À Susana AGRADECIMENTOS Esta tese é o resultado de um trabalho conjunto para o qual muitos contribuíram e aos quais desejo reconhecer e deixar o meu agradecimento. Ao professor Doutor José Paulo Sousa, meu orientador, pela indispensável ajuda, paciência e orientação científica. Ao professor Doutor José Manuel Jesus, meu orientador, pela amizade e apoio desde os primeiros momentos. Pelo seu empenho, conselhos transmitidos, chamadas à razão e orientação científica o meu muito obrigado. Ao Doutor Pedro Cardoso, meu orientador e a quem muito devo, pelo constante acompanhamento e disponibilidade, amizade e orientação científica. Por tudo o que me ensinou, pela motivação e animo que sempre me transmitiu, e, acima de tudo, pela manutenção da objectividade do trabalho. -

Succineidae, Testacelloidea and Helicoidea
Zootaxa 3721 (2): 157–171 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2013 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3721.2.3 http://zoobank.org/urn:lsid:zoobank.org:pub:71B4B001-FB10-4B99-ACF9-720131457534 The fossil pulmonate snails of Sandelzhausen (Early/Middle Miocene, Germany): Succineidae, Testacelloidea and Helicoidea RODRIGO BRINCALEPE SALVADOR Staatliches Museum für Naturkunde Stuttgart (Stuttgart, Germany). Mathematisch-Naturwissenschaftliche Fakultät, Eberhard Karls Universität Tübingen (Tübingen, Germany). E-mail: [email protected] Abstract Sandelzhausen is an Early/Middle Miocene (Mammal Neogene zone MN5) fossil site near Mainburg, S Germany, and despite its small size it harbors a rich fossil record. Hundreds of fossil continental mollusks, almost exclusively pulmo- nates snails, were recovered during the excavations, but never received due attention by researchers. Here, the second part of a formal taxonomical treatment of the fossil pulmonates from Sandelzhausen is presented, dealing with the superfam- ilies Succineoidea, Testacelloidea and Helicoidea, and including the description of a new hygromiid species. The follow- ing species were found in the material: Succinea minima (Succineidae); Palaeoglandina sp. (Spiraxidae); Testacella zellii (Testacellidae); Klikia cf. coarctata (Elonidae); Cepaea cf. eversa, Cepaea cf. sylvestrina and Tropidomphalus cf. incras- satus (Helicidae); ?Helicodonta sp. and Helicodontidae indet. (Helicodontidae); Leucochroopsis kleinii and Urticicola perchtae sp. nov. (Hygromiidae). Key words: Gastropoda, MN5 European Mammal Neogene zone, Pulmonata, Stylommatophora, Urticicola perchtae new species Introduction The Sandelzhausen fossil site is one of the most important continental sites in Europe (Moser et al. 2009a) and its bounty include hundreds of specimens of gastropods. -

Malaco Le Journal Électronique De La Malacologie Continentale Française
MalaCo Le journal électronique de la malacologie continentale française www.journal-malaco.fr MalaCo (ISSN 1778-3941) est un journal électronique gratuit, annuel ou bisannuel pour la promotion et la connaissance des mollusques continentaux de la faune de France. Equipe éditoriale Jean-Michel BICHAIN / Paris / [email protected] Xavier CUCHERAT / Audinghen / [email protected] Benoît FONTAINE / Paris / [email protected] Olivier GARGOMINY / Paris / [email protected] Vincent PRIE / Montpellier / [email protected] Les manuscrits sont à envoyer à : Journal MalaCo Muséum national d’Histoire naturelle Equipe de Malacologie Case Postale 051 55, rue Buffon 75005 Paris Ou par Email à [email protected] MalaCo est téléchargeable gratuitement sur le site : http://www.journal-malaco.fr MalaCo (ISSN 1778-3941) est une publication de l’association Caracol Association Caracol Route de Lodève 34700 Saint-Etienne-de-Gourgas JO Association n° 0034 DE 2003 Déclaration en date du 17 juillet 2003 sous le n° 2569 Journal électronique de la malacologie continentale française MalaCo Septembre 2006 ▪ numéro 3 Au total, 119 espèces et sous-espèces de mollusques, dont quatre strictement endémiques, sont recensées dans les différents habitats du Parc naturel du Mercantour (photos Olivier Gargominy, se reporter aux figures 5, 10 et 17 de l’article d’O. Gargominy & Th. Ripken). Sommaire Page 100 Éditorial Page 101 Actualités Page 102 Librairie Page 103 Brèves & News ▪ Endémisme et extinctions : systématique des Endodontidae (Mollusca, Pulmonata) de Rurutu (Iles Australes, Polynésie française) Gabrielle ZIMMERMANN ▪ The first annual meeting of Task-Force-Limax, Bünder Naturmuseum, Chur, Switzerland, 8-10 September, 2006: presentation, outcomes and abstracts Isabel HYMAN ▪ Collecting and transporting living slugs (Pulmonata: Limacidae) Isabel HYMAN ▪ A List of type specimens of land and freshwater molluscs from France present in the national molluscs collection of the Hebrew University of Jerusalem Henk K. -

Molluscs of the Dürrenstein Wilderness Area
Molluscs of the Dürrenstein Wilderness Area S a b i n e F ISCHER & M i c h a e l D UDA Abstract: Research in the Dürrenstein Wilderness Area (DWA) in the southwest of Lower Austria is mainly concerned with the inventory of flora, fauna and habitats, interdisciplinary monitoring and studies on ecological disturbances and process dynamics. During a four-year qualitative study of non-marine molluscs, 96 sites within the DWA and nearby nature reserves were sampled in cooperation with the “Alpine Land Snails Working Group” located at the Natural History Museum of Vienna. Altogether, 84 taxa were recorded (72 land snails, 12 water snails and mussels) including four endemics and seven species listed in the Austrian Red List of Molluscs. A reference collection (empty shells) of molluscs, which is stored at the DWA administration, was created. This project was the first systematic survey of mollusc fauna in the DWA. Further sampling might provide additional information in the future, particularly for Hydrobiidae in springs and caves, where detailed analyses (e.g. anatomical and genetic) are needed. Key words: Wilderness Dürrenstein, Primeval forest, Benign neglect, Non-intervention management, Mollusca, Snails, Alpine endemics. Introduction manifold species living in the wilderness area – many of them “refugees”, whose natural habitats have almost In concordance with the IUCN guidelines, research is disappeared in today’s over-cultivated landscape. mandatory for category I wilderness areas. However, it may not disturb the natural habitats and communities of the nature reserve. Research in the Dürrenstein The Dürrenstein Wilderness Area Wilderness Area (DWA) focuses on providing invento- (DWA) ries of flora and fauna, on interdisciplinary monitoring The Dürrenstein Wilderness Area (DWA) was as well as on ecological disturbances and process dynamics. -

Fauna of New Zealand Ko Te Aitanga Pepeke O Aotearoa
aua o ew eaa Ko te Aiaga eeke o Aoeaoa IEEAE SYSEMAICS AISOY GOU EESEAIES O ACAE ESEAC ema acae eseac ico Agicuue & Sciece Cee P O o 9 ico ew eaa K Cosy a M-C aiièe acae eseac Mou Ae eseac Cee iae ag 917 Aucka ew eaa EESEAIE O UIESIIES M Emeso eame o Eomoogy & Aima Ecoogy PO o ico Uiesiy ew eaa EESEAIE O MUSEUMS M ama aua Eiome eame Museum o ew eaa e aa ogaewa O o 7 Weigo ew eaa EESEAIE O OESEAS ISIUIOS awece CSIO iisio o Eomoogy GO o 17 Caea Ciy AC 1 Ausaia SEIES EIO AUA O EW EAA M C ua (ecease ue 199 acae eseac Mou Ae eseac Cee iae ag 917 Aucka ew eaa Fauna of New Zealand Ko te Aitanga Pepeke o Aotearoa Number / Nama 38 Naturalised terrestrial Stylommatophora (Mousca Gasooa Gay M ake acae eseac iae ag 317 amio ew eaa 4 Maaaki Whenua Ρ Ε S S ico Caeuy ew eaa 1999 Coyig © acae eseac ew eaa 1999 o a o is wok coee y coyig may e eouce o coie i ay om o y ay meas (gaic eecoic o mecaica icuig oocoyig ecoig aig iomaio eiea sysems o oewise wiou e wie emissio o e uise Caaoguig i uicaio AKE G Μ (Gay Micae 195— auase eesia Syommaooa (Mousca Gasooa / G Μ ake — ico Caeuy Maaaki Weua ess 1999 (aua o ew eaa ISS 111-533 ; o 3 IS -7-93-5 I ie 11 Seies UC 593(931 eae o uIicaio y e seies eio (a comee y eo Cosy usig comue-ase e ocessig ayou scaig a iig a acae eseac M Ae eseac Cee iae ag 917 Aucka ew eaa Māoi summay e y aco uaau Cosuas Weigo uise y Maaaki Weua ess acae eseac O o ico Caeuy Wesie //wwwmwessco/ ie y G i Weigo o coe eoceas eicuaum (ue a eigo oaa (owe (IIusao G M ake oucio o e coou Iaes was ue y e ew eaIa oey oa ue oeies eseac -
Ecological Groups of Snails – Use and Perspectives
The subdivision of all central European Holocene and Late Glacial land snail species to ecological groups ecological Glacial Early Holocene Middle Holocene Late Holocene (sensu Walker at al 2012) modern immigrants comment group Acanthinula aculeata Acanthinula aculeata Acanthinula aculeata Acanthinula aculeata Acicula parcelineata Acicula parcelineata Aegopinella epipedostoma one sites Aegopinella nitens Aegopinella nitens Aegopinella nitidula Aegopinella nitidula few sites Aegopinella pura Aegopinella pura Aegopinella pura Aegopinella pura Aegopis verticillus Ecological groups of snails Argna bielzi Argna bielzi Bulgarica cana Bulgarica cana Carpathica calophana Carpathica calophana one site; undated Causa holosericea Causa holosericea Clausilia bidentata no fossil data Clausilia cruciata Clausilia cruciata Clausilia cruciata – use and perspectives Cochlodina laminata Cochlodina laminata Cochlodina laminata Cochlodina laminata Cochlodina orthostoma Cochlodina orthostoma Cochlodina orthostoma Cochlodina orthostoma Daudebardia brevipes Daudebardia brevipes Daudebardia rufa Daudebardia rufa Daudebardia rufa Daudebardia rufa Discus perspectivus Discus perspectivus Discus perspectivus 1 2 1 1 ) Lucie Juřičková , Michal Horsák , Jitka Horáčková and Vojen Ložek Discus ruderatus Discus ruderatus Discus ruderatus Discus ruderatus Ena montana Ena montana Ena montana Ena montana forest Eucobresia nivalis Eucobresia nivalis Eucobresia nivalis Faustina faustina Faustina faustina Faustina faustina Faustina faustina Faustina rossmaessleri Faustina -

293 – Les Noms Scientifiques Français Des Mollusques Continentaux De
LES NOMS SCIENTIFIQUES FRANÇAIS DES MOLLUSQUES CONTINENTAUX DE FRANCE : processus d’ÉtABLISSEMENT d’une LISTE DE RÉFÉRENCE Benoît FONTAINE 1, Jean-Michel BICHAIN 2, Xavier CUCHERAT 3, Olivier GAR G OMINY 4 & Vincent PRIÉ 5 SUMMARY. — French scientific names of continental molluscs of France: process for establishing a list of reference. — In the biodiversity crisis context and with the increasing general awareness on this issue, conservation of small and poorly-known species is hampered by the fact they only have latine names. In order to communicate for biodiversity conservation, having French names is an advantage which is lacking in terrestrial and freshwater molluscs from France. To remedy this problem, we propose a list of French scientific names for this group, i.e. all species and subspecies known from France. We have listed existing names in legal documents, in usage and in the 18th and 19th centuries scientific literature. The resulting list being incomplete, we had to create new French names, following a series of recommendations adapted from similar works dealing with other taxonomic groups. We conclude by dealing with the issue of the legitimacy and validity of such names. The list of French scientific names is given as an appendix and is downloadable from internet. RÉSUMÉ. — Dans le contexte de la crise de la biodiversité et de la prise de conscience par le grand public des enjeux environnementaux, la conservation des espèces petites et méconnues est handicapée par le fait que ces espèces ne peuvent être désignées que par leur nom latin. Dans une optique de communication pour la préservation de la biodiversité, disposer de nom français est un atout qui fait défaut pour les mol- lusques terrestres et d’eau douce de France. -

(Approx) Mixed Micro Shells (22G Bags) Philippines € 10,00 £8,64 $11,69 Each 22G Bag Provides Hours of Fun; Some Interesting Foraminifera Also Included
Special Price £ US$ Family Genus, species Country Quality Size Remarks w/o Photo Date added Category characteristic (€) (approx) (approx) Mixed micro shells (22g bags) Philippines € 10,00 £8,64 $11,69 Each 22g bag provides hours of fun; some interesting Foraminifera also included. 17/06/21 Mixed micro shells Ischnochitonidae Callistochiton pulchrior Panama F+++ 89mm € 1,80 £1,55 $2,10 21/12/16 Polyplacophora Ischnochitonidae Chaetopleura lurida Panama F+++ 2022mm € 3,00 £2,59 $3,51 Hairy girdles, beautifully preserved. Web 24/12/16 Polyplacophora Ischnochitonidae Ischnochiton textilis South Africa F+++ 30mm+ € 4,00 £3,45 $4,68 30/04/21 Polyplacophora Ischnochitonidae Ischnochiton textilis South Africa F+++ 27.9mm € 2,80 £2,42 $3,27 30/04/21 Polyplacophora Ischnochitonidae Stenoplax limaciformis Panama F+++ 16mm+ € 6,50 £5,61 $7,60 Uncommon. 24/12/16 Polyplacophora Chitonidae Acanthopleura gemmata Philippines F+++ 25mm+ € 2,50 £2,16 $2,92 Hairy margins, beautifully preserved. 04/08/17 Polyplacophora Chitonidae Acanthopleura gemmata Australia F+++ 25mm+ € 2,60 £2,25 $3,04 02/06/18 Polyplacophora Chitonidae Acanthopleura granulata Panama F+++ 41mm+ € 4,00 £3,45 $4,68 West Indian 'fuzzy' chiton. Web 24/12/16 Polyplacophora Chitonidae Acanthopleura granulata Panama F+++ 32mm+ € 3,00 £2,59 $3,51 West Indian 'fuzzy' chiton. 24/12/16 Polyplacophora Chitonidae Chiton tuberculatus Panama F+++ 44mm+ € 5,00 £4,32 $5,85 Caribbean. 24/12/16 Polyplacophora Chitonidae Chiton tuberculatus Panama F++ 35mm € 2,50 £2,16 $2,92 Caribbean. 24/12/16 Polyplacophora Chitonidae Chiton tuberculatus Panama F+++ 29mm+ € 3,00 £2,59 $3,51 Caribbean. -

Why Do Snails Have Hairs? a Bayesian Inference of Character Evolution Markus Pfenninger*1, Magda Hrabáková2, Dirk Steinke3 and Aline Dèpraz4
BMC Evolutionary Biology BioMed Central Research article Open Access Why do snails have hairs? A Bayesian inference of character evolution Markus Pfenninger*1, Magda Hrabáková2, Dirk Steinke3 and Aline Dèpraz4 Address: 1Abteilung Ökologie & Evolution, J.W. Goethe-Universität, BioCampus Siesmayerstraße, 60054 Frankfurt/Main, Germany, 2Deparment of Zoology, Charles University, Viniènà 7, 128 44 Praha 2, Czech Republic, 3Department of Biology, University of Konstanz, Postbox 5560 M618, 78457 Konstanz, Germany and 4Département d'Ecologie et Evolution, Université de Lausanne, Bâtiment de Biologie, Dorigny, 1015 Lausanne, Switzerland Email: Markus Pfenninger* - [email protected]; Magda Hrabáková - [email protected]; Dirk Steinke - [email protected]; Aline Dèpraz - [email protected] * Corresponding author Published: 04 November 2005 Received: 14 July 2005 Accepted: 04 November 2005 BMC Evolutionary Biology 2005, 5:59 doi:10.1186/1471-2148-5-59 This article is available from: http://www.biomedcentral.com/1471-2148/5/59 © 2005 Pfenninger et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Costly structures need to represent an adaptive advantage in order to be maintained over evolutionary times. Contrary to many other conspicuous shell ornamentations of gastropods, the haired shells of several Stylommatophoran land snails still lack a convincing adaptive explanation. In the present study, we analysed the correlation between the presence/absence of hairs and habitat conditions in the genus Trochulus in a Bayesian framework of character evolution. -
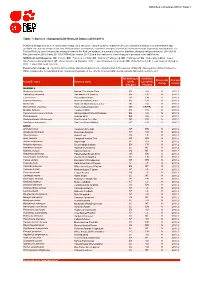
Table 7: Species Changing IUCN Red List Status (2010-2011)
IUCN Red List version 2011.2: Table 7 Table 7: Species changing IUCN Red List Status (2010-2011) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2010 (IUCN Red List version 2010.4) and 2011 (IUCN Red List version 2011.2) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.) IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2010) List (2011) change version Category Category MAMMALS Bradypus torquatus Maned Three-toed Sloth EN VU N 2011.1 Callicebus oenanthe San Martin Titi Monkey EN CR N 2011.1 Equus ferus Przewalski's Horse CR EN G 2011.2 -

Fossil Flora and Fauna of Bosnia and Herzegovina D Ela
FOSSIL FLORA AND FAUNA OF BOSNIA AND HERZEGOVINA D ELA Odjeljenje tehničkih nauka Knjiga 10/1 FOSILNA FLORA I FAUNA BOSNE I HERCEGOVINE Ivan Soklić DOI: 10.5644/D2019.89 MONOGRAPHS VOLUME LXXXIX Department of Technical Sciences Volume 10/1 FOSSIL FLORA AND FAUNA OF BOSNIA AND HERZEGOVINA Ivan Soklić Ivan Soklić – Fossil Flora and Fauna of Bosnia and Herzegovina Original title: Fosilna flora i fauna Bosne i Hercegovine, Sarajevo, Akademija nauka i umjetnosti Bosne i Hercegovine, 2001. Publisher Academy of Sciences and Arts of Bosnia and Herzegovina For the Publisher Academician Miloš Trifković Reviewers Dragoljub B. Đorđević Ivan Markešić Editor Enver Mandžić Translation Amra Gadžo Proofreading Amra Gadžo Correction Sabina Vejzagić DTP Zoran Buletić Print Dobra knjiga Sarajevo Circulation 200 Sarajevo 2019 CIP - Katalogizacija u publikaciji Nacionalna i univerzitetska biblioteka Bosne i Hercegovine, Sarajevo 57.07(497.6) SOKLIĆ, Ivan Fossil flora and fauna of Bosnia and Herzegovina / Ivan Soklić ; [translation Amra Gadžo]. - Sarajevo : Academy of Sciences and Arts of Bosnia and Herzegovina = Akademija nauka i umjetnosti Bosne i Hercegovine, 2019. - 861 str. : ilustr. ; 25 cm. - (Monographs / Academy of Sciences and Arts of Bosnia and Herzegovina ; vol. 89. Department of Technical Sciences ; vol. 10/1) Prijevod djela: Fosilna flora i fauna Bosne i Hercegovine. - Na spor. nasl. str.: Fosilna flora i fauna Bosne i Hercegovine. - Bibliografija: str. 711-740. - Registri. ISBN 9958-501-11-2 COBISS/BIH-ID 8839174 CONTENTS FOREWORD ........................................................................................................... -

Malaco 04 Full Issue 2007.Pdf
MalaCo Le journal électronique de la malacologie continentale française www.journal-malaco.fr MalaCo (ISSN 1778-3941) est un journal électronique gratuit, annuel ou bisannuel pour la promotion et la connaissance des mollusques continentaux de la faune de France. Equipe éditoriale Jean-Michel BICHAIN / Paris / [email protected] Xavier CUCHERAT / Audinghen / [email protected] (Editeur en chef du numéro 4) Benoît FONTAINE / Paris / [email protected] Olivier GARGOMINY / Paris / [email protected] Vincent PRIE / Montpellier / [email protected] Collaborateurs de ce numéro Gilbert COCHET Robert COWIE Sylvain DEMUYNCK Daniel PAVON Sylvain VRIGNAUD Pour soumettre un article à MalaCo : 1ère étape – Le premier auteur veillera à ce que le manuscrit soit conforme aux recommandations aux auteurs (en fin de ce numéro ou consultez le site www.journal-malaco.fr). Dans le cas contraire, la rédaction peut se réserver le droit de refuser l’article. 2ème étape – Joindre une lettre à l’éditeur, en document texte, en suivant le modèle suivant : "Veuillez trouvez en pièce jointe l’article rédigé par << mettre les noms et prénoms de tous les auteurs>> et intitulé : << mettre le titre en français et en anglais >> (avec X pages, X figures et X tableaux). Les auteurs cèdent au journal MalaCo (ISSN1778-3941) le droit de publication de ce manuscrit et ils garantissent que l’article est original, qu’il n’a pas été soumis pour publication à un autre journal, n’a pas été publié auparavant et que tous sont en accord avec le contenu." 3ème étape – Envoyez par voie électronique le manuscrit complet (texte et figures) en format .doc et la lettre à l’éditeur à : [email protected].