Enhanced Recovery of CD9-Positive Extracellular Vesicles from Human Specimens by Chelating Reagent
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Common Gene Polymorphisms Associated with Thrombophilia
Chapter 5 Common Gene Polymorphisms Associated with Thrombophilia Christos Yapijakis, Zoe Serefoglou and Constantinos Voumvourakis Additional information is available at the end of the chapter http://dx.doi.org/10.5772/61859 Abstract Genetic association studies have revealed a correlation between DNA variations in genes encoding factors of the hemostatic system and thrombosis-related disease. Certain var‐ iant alleles of these genes that affect either gene expression or function of encoded protein are known to be genetic risk factors for thrombophilia. The chapter presents the current genetics and molecular biology knowledge of the most important DNA polymorphisms in thrombosis-related genes encoding coagulation factor V (FV), coagulation factor II (FII), coagulation factor XII (FXII), coagulation factor XIII A1 subunit (FXIIIA1), 5,10- methylene tetrahydrofolate reductase (MTHFR), serpine1 (SERPINE1), angiotensin I-con‐ verting enzyme (ACE), angiotensinogen (AGT), integrin A2 (ITGA2), plasma carboxypeptidase B2 (CPB2), platelet glycoprotein Ib α polypeptide (GP1BA), thrombo‐ modulin (THBD) and protein Z (PROZ). The molecular detection methods of each DNA polymorphism is presented, in addition to the current knowledge regarding its influence on thrombophilia and related thrombotic events, including stroke, myocardial infarction, deep vein thrombosis, spontaneous abortion, etc. In addition, best thrombosis prevention strategies with a combination of genetic counseling and molecular testing are discussed. Keywords: Thrombophilia, coagulation -

A Differential Protein Solubility Approach for the Depletion of Highly Abundant Proteins in Plasma Using Ammonium Sulfate Ravi Chand Bollineni1, 2*, Ingrid J
Electronic Supplementary Material (ESI) for Analyst. This journal is © The Royal Society of Chemistry 2015 A differential protein solubility approach for the depletion of highly abundant proteins in plasma using ammonium sulfate Ravi Chand Bollineni1, 2*, Ingrid J. Guldvik3, Henrik Gronberg4, Fredrik Wiklund4, Ian G. Mills3, 5, 6 and Bernd Thiede1, 2 1Department of Biosciences, University of Oslo, Oslo, Norway 2Biotechnology Centre of Oslo, University of Oslo, Oslo, Norway 3Centre for Molecular Medicine Norway (NCMM), University of Oslo and Oslo University Hospitals, Norway 4Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm, Sweden 5Department of Cancer Prevention, Oslo University Hospitals, Oslo, Norway 6Department of Urology, Oslo University Hospitals, Oslo, Norway Keywords: ammonium sulfate, blood, depletion, plasma, protein precipitation *To whom the correspondence should be addressed: Ravi Chand Bollineni, Department of Biosciences, University of Oslo, P.O. Box 1066 Blindern, 0316 Oslo, Norway, Tel.: +47-22840512; Fax +47-22840501; E-mail: [email protected] Supplementary information Figure S1: SDS-PAGE analysis of serum proteins precipitated with the ethanol/sodium acetate (A), TCA/acetone (B) and ammonium sulfate precipitation (C). A) Ethanol/sodium acetate precipitation: (1) pellet obtained after 42% ethanol precipitation and (2) pellet obtained after precipitation of proteins in the supernatant with 0.8M sodium acetate (pH5.7) and (3) proteins left over in the supernatant. B) Serum proteins are precipitated with 10% TCA/acetone (1) and (2) proteins left over in the supernatant. C) Serum proteins are precipitated with increasing ammonium sulfate concentrations 15% (1), 25% (2), 35% (3), 40% (4), 45% (5), 50% (6) and total serum (7). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
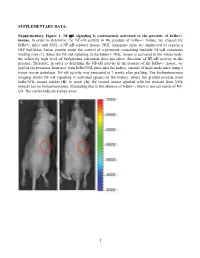
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

Carboxypeptidase B2 (CPB2) Human Shrna Lentiviral Particle (Locus ID 1361) Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TL305252V Carboxypeptidase B2 (CPB2) Human shRNA Lentiviral Particle (Locus ID 1361) Product data: Product Type: shRNA Lentiviral Particles Product Name: Carboxypeptidase B2 (CPB2) Human shRNA Lentiviral Particle (Locus ID 1361) Locus ID: 1361 Synonyms: CPU; PCPB; TAFI Vector: pGFP-C-shLenti (TR30023) Format: Lentiviral particles RefSeq: NM_001278541, NM_001872, NM_016413, NM_001872.1, NM_001872.2, NM_001872.3, NM_001872.4, NM_016413.1, NM_016413.2, NM_016413.3, NM_001278541.1, BC007057, BC007057.1, NM_001872.5 Summary: Carboxypeptidases are enzymes that hydrolyze C-terminal peptide bonds. The carboxypeptidase family includes metallo-, serine, and cysteine carboxypeptidases. According to their substrate specificity, these enzymes are referred to as carboxypeptidase A (cleaving aliphatic residues) or carboxypeptidase B (cleaving basic amino residues). The protein encoded by this gene is activated by trypsin and acts on carboxypeptidase B substrates. After thrombin activation, the mature protein downregulates fibrinolysis. Polymorphisms have been described for this gene and its promoter region. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jun 2013] shRNA Design: These shRNA constructs were designed against multiple splice variants at this gene locus. To be certain that your variant of interest is targeted, -

Electronic Supplementary Material (ESI) for Analyst. This Journal Is © the Royal Society of Chemistry 2020
Electronic Supplementary Material (ESI) for Analyst. This journal is © The Royal Society of Chemistry 2020 Table S1. -

Genetic Factors for Obesity
843-851 5/10/06 13:36 Page 843 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 18: 843-851, 2006 843 Genetic factors for obesity YOSHIJI YAMADA1,2, KIMIHIKO KATO3, TAKASHI KAMEYAMA4, KIYOSHI YOKOI3, HITOSHI MATSUO5, TOMONORI SEGAWA5, SACHIRO WATANABE5, SAHOKO ICHIHARA1, HIDEMI YOSHIDA6, KEI SATOH6 and YOSHINORI NOZAWA2 1Department of Human Functional Genomics, Life Science Research Center, Mie University, Tsu; 2Gifu International Institute of Biotechnology, Kakamigahara; Departments of 3Cardiovascular Medicine and 4Neurology, Gifu Prefectural Tajimi Hospital, Tajimi; 5Department of Cardiology, Gifu Prefectural Gifu Hospital, Gifu; 6Department of Vascular Biology, Institute of Brain Science, Hirosaki University School of Medicine, Hirosaki, Japan Received May 29, 2006; Accepted July 20, 2006 Abstract. The purpose of the present study was to identify Introduction gene polymorphisms for the reliable assessment of genetic factors for obesity. The study population comprised 3906 The prevalence of obesity, a multifactorial disease caused by unrelated Japanese individuals (2286 men, 1620 women), an interaction of genetic factors with lifestyle and environ- including 1196 subjects (677 men, 519 women) with obesity mental factors (1), is rapidly increasing worldwide. A sedentary (body mass index of ≥25 kg/m2) and 2710 controls (1609 men, lifestyle, high-fat and high-energy diet, and genetic pre- 1101 women). The genotypes for 147 polymorphisms of 124 disposition to obesity all contribute to the epidemic. Although candidate genes were determined with a method that combines genetic linkage analyses (2-5) and candidate gene approaches the polymerase chain reaction and sequence-specific (6-9) have implicated several loci and candidate genes in oligonucleotide probes with suspension array technology. predisposition to obesity, the genes that contribute to genetic Multivariable logistic regression analysis with adjustment for susceptibility to this condition remain to be identified age, sex, and the prevalence of smoking revealed that the - definitively. -

Common Gene Polymorphisms Associated with Thrombophilia
Chapter 5 Common Gene Polymorphisms Associated with Thrombophilia Christos Yapijakis, Zoe Serefoglou and Constantinos Voumvourakis Additional information is available at the end of the chapter http://dx.doi.org/10.5772/61859 Abstract Genetic association studies have revealed a correlation between DNA variations in genes encoding factors of the hemostatic system and thrombosis-related disease. Certain var‐ iant alleles of these genes that affect either gene expression or function of encoded protein are known to be genetic risk factors for thrombophilia. The chapter presents the current genetics and molecular biology knowledge of the most important DNA polymorphisms in thrombosis-related genes encoding coagulation factor V (FV), coagulation factor II (FII), coagulation factor XII (FXII), coagulation factor XIII A1 subunit (FXIIIA1), 5,10- methylene tetrahydrofolate reductase (MTHFR), serpine1 (SERPINE1), angiotensin I-con‐ verting enzyme (ACE), angiotensinogen (AGT), integrin A2 (ITGA2), plasma carboxypeptidase B2 (CPB2), platelet glycoprotein Ib α polypeptide (GP1BA), thrombo‐ modulin (THBD) and protein Z (PROZ). The molecular detection methods of each DNA polymorphism is presented, in addition to the current knowledge regarding its influence on thrombophilia and related thrombotic events, including stroke, myocardial infarction, deep vein thrombosis, spontaneous abortion, etc. In addition, best thrombosis prevention strategies with a combination of genetic counseling and molecular testing are discussed. Keywords: Thrombophilia, coagulation -

Proteomics Analysis Reveals a Th17-Prone Cell Population in Pre-Symptomatic Graft- Versus-Host Disease
Supplemental Data for: Proteomics analysis reveals a Th17-prone cell population in pre-symptomatic graft- versus-host disease Authors: Wei Li,1* Liangyi Liu,1* Aurelie Gomez,2* Jilu Zhang,1 Abdulraouf Ramadan,1 Qing Zhang,3 Sung W. Choi,2 Peng Zhang,2 Joel K. Greenson,2 Chen Liu,4 Di Jiang,5 Elizabeth Virts,1 Stephanie L. Kelich,1 Hong Wei Chu,5 Ryan Flynn,6 Bruce R. Blazar,6 Helmut Hanenberg,1 Samir Hanash,7 and Sophie Paczesny1‡ Affiliations: 1Indiana University School of Medicine, Indianapolis, IN, USA; 2University of Michigan, Ann Arbor, MI, USA; 3Fred Hutchinson Cancer Research Center, Seattle, WA, USA; 4Rutgers-Robert Wood Johnson Medical School, New Brunswick, NJ, USA; 5National Jewish Health, Denver, CO, USA; 6University of Minnesota, Minneapolis, MN, USA; 7MD Anderson Cancer Center, Houston, TX, USA. *These authors contributed equally to this work. ‡Corresponding author. E-mail: [email protected] This PDF file includes: Supplemental Figure 1. Conventional T cells (Tcons) and Tregs gating strategy. Supplemental Figure 2. Absolute lymphocyte counts and correlation of CD4+CD146+CCR5+ T-cell counts and frequencies of CD4+CD146+CCR5+ T cells in PB of HCT patients. Supplemental Figure 3. Frequencies of CD4+CD146+ T cells and CD4+CCR5+ T cells in PB of HCT patients. Supplemental Figure 4. CD146 expression on CD4 and CD8 T cells in PB of healthy donors and GI-GVHD patients. Supplemental Figure 5. Receiver operating characteristic (ROC) curve comparing GI-GVHD versus non-GVHD enteritis HCT patients. Supplemental Figure 6. CD4+CD146+CCR5+ T-cell frequency stratified by maximum GI- GVHD stage. -

Both Full-Length and Protease-Cleaved Products of Osteopontin Are Elevated in Infectious Diseases
biomedicines Review Both Full-Length and Protease-Cleaved Products of Osteopontin Are Elevated in Infectious Diseases Toshio Hattori 1,* , Hiroko Iwasaki-Hozumi 1 , Gaowa Bai 1 , Haorile Chagan-Yasutan 1,2, Ashwnini Shete 3 , Elizabeth Freda Telan 4, Atsushi Takahashi 1, Yugo Ashino 5 and Takashi Matsuba 6 1 Research Institute of Health and Welfare, Kibi International University, Takahashi 716-8508, Japan; [email protected] (H.I.-H.); [email protected] (G.B.); [email protected] (H.C.-Y.); [email protected] (A.T.) 2 Mongolian Psychosomatic Medicine Department, International Mongolian Medicine Hospital of Inner Mongolia, Hohhot 010065, China 3 ICMR-National AIDS Research Institute, 73 G-Block, MIDC, Bhosari, Pune 411026, India; [email protected] 4 STD AIDS Cooperative Central Laboratory, San Lazaro Hospital, Manila 1003, Philippines; [email protected] 5 Department of Respiratory Medicine, Sendai City Hospital, Sendai 982-8502, Japan; [email protected] 6 Department of Animal Pharmaceutical Science, School of Pharmaceutical Science, Kyusyu University of Health and Welfare, Nobeoka 882-8508, Japan; [email protected] * Correspondence: [email protected]; Tel./Fax: +81-866-22-9469 Citation: Hattori, T.; Abstract: Circulating full-length osteopontin (FL-OPN) is elevated in plasma from patients with Iwasaki-Hozumi, H.; Bai, G.; various infectious diseases, such as adult T-cell leukemia, Mycobacterium tuberculosis (TB), hepatitis Chagan-Yasutan, H.; Shete, A.; Telan, virus infection, leptospirosis, acquired immune deficiency syndrome (AIDS), AIDS/TB, and coron- E.F.; Takahashi, A.; Ashino, Y.; avirus disease 2019 (COVID-19). Proteolysis of OPN by thrombin, matrix metalloproteases, caspase Matsuba, T. -

High Fat Diet Delays Plasmin Generation in a Thrombomodulin-Dependent Manner in Mice
American Society of Hematology 2021 L Street NW, Suite 900, Washington, DC 20036 Phone: 202-776-0544 | Fax 202-776-0545 [email protected] High fat diet delays plasmin generation in a thrombomodulin-dependent manner in mice Downloaded from https://ashpublications.org/blood/article-pdf/doi/10.1182/blood.2019004267/1716422/blood.2019004267.pdf by UNIV OF NC/ACQ SRVCS user on 25 March 2020 Tracking no: BLD-2019-004267R1 Adam Miszta (University of North Carolina at Chapel Hill, United States) Anna Kopec (University of North Carolina at Chapel Hill, United States) Asmita Pant (Michigan State University, ) Lori Holle (University of North Carolina, United States) James Byrnes (University of California, San Francisco, United States) Daniel Lawrence (University of Michigan Medical School, United States) Kirk Hansen (UC Denver, United States) Matthew Flick (University of North Carolina, Chapel Hill, United States) James Luyendyk (Michigan State University, United States) Bas de Laat (Synapse BV, Netherlands) Alisa Wolberg (University of North Carolina at Chapel Hill, United States) Abstract: Obesity is a prevalent prothrombotic risk factor marked by enhanced fibrin formation and suppressed fibrinolysis. Fibrin both promotes thrombotic events and drives obesity pathophysiology, but a lack of essential analytical tools has left fibrinolytic mechanisms affected by obesity poorly defined. Using a plasmin-specific fluorogenic substrate, we developed a plasmin generation (PG) assay that is sensitive to tissue plasminogen activator, α2-antiplasmin, active plasminogen activator inhibitor (PAI-1), and fibrin formation, but not fibrin crosslinking in mouse plasma. Compared to plasma from mice fed a control diet (CD), plasmas from mice fed a high fat diet (HFD) showed delayed PG and reduced PG velocity. -

COVID-19 Biomarkers for Severity Mapped to Polycystic Ovary Syndrome
Moin et al. J Transl Med (2020) 18:490 https://doi.org/10.1186/s12967-020-02669-2 Journal of Translational Medicine LETTER TO THE EDITOR Open Access COVID-19 biomarkers for severity mapped to polycystic ovary syndrome Abu Saleh Md Moin1, Thozhukat Sathyapalan2, Stephen L. Atkin3† and Alexandra E. Butler1*† To the Editor, diagnostic criteria. Proteins that were identifed as being altered in COVID-19 disease for vessel damage (16 pro- Large scale multi-omics analysis has identifed signif- teins), platelet degranulation (11 proteins), coagula- cant diferences in the biomarkers between COVID-19 tion cascade (24 proteins) and acute phase response (9 disease and control subjects [1]. Tese protein panels tar- proteins), shown in Table 1, were determined by Slow get biological processes involved in vessel damage, plate- Of-rate Modifed Aptamer (SOMA)-scan plasma pro- let degranulation, the coagulation cascade and the acute tein measurement [6]. Statistics were performed using phase response [1], with greater protein changes depend- Graphpad Prism 8.0. ent on the COVID-19 severity. However, it is observed As reported previously [2], cohorts were age-matched, that in metabolic conditions such as polycystic ovary but PCOS women had increased insulin resistance, syndrome expressed proteins difer compared to control androgens and CRP (p < 0.001): systolic and diastolic women [2] and PCOS patients have increased platelet blood pressure, and waist circumference were higher aggregation and decreased plasma fbrinolytic activity, (p < 0.05). resulting in