Genetic Factors for Obesity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Common Gene Polymorphisms Associated with Thrombophilia
Chapter 5 Common Gene Polymorphisms Associated with Thrombophilia Christos Yapijakis, Zoe Serefoglou and Constantinos Voumvourakis Additional information is available at the end of the chapter http://dx.doi.org/10.5772/61859 Abstract Genetic association studies have revealed a correlation between DNA variations in genes encoding factors of the hemostatic system and thrombosis-related disease. Certain var‐ iant alleles of these genes that affect either gene expression or function of encoded protein are known to be genetic risk factors for thrombophilia. The chapter presents the current genetics and molecular biology knowledge of the most important DNA polymorphisms in thrombosis-related genes encoding coagulation factor V (FV), coagulation factor II (FII), coagulation factor XII (FXII), coagulation factor XIII A1 subunit (FXIIIA1), 5,10- methylene tetrahydrofolate reductase (MTHFR), serpine1 (SERPINE1), angiotensin I-con‐ verting enzyme (ACE), angiotensinogen (AGT), integrin A2 (ITGA2), plasma carboxypeptidase B2 (CPB2), platelet glycoprotein Ib α polypeptide (GP1BA), thrombo‐ modulin (THBD) and protein Z (PROZ). The molecular detection methods of each DNA polymorphism is presented, in addition to the current knowledge regarding its influence on thrombophilia and related thrombotic events, including stroke, myocardial infarction, deep vein thrombosis, spontaneous abortion, etc. In addition, best thrombosis prevention strategies with a combination of genetic counseling and molecular testing are discussed. Keywords: Thrombophilia, coagulation -

A Differential Protein Solubility Approach for the Depletion of Highly Abundant Proteins in Plasma Using Ammonium Sulfate Ravi Chand Bollineni1, 2*, Ingrid J
Electronic Supplementary Material (ESI) for Analyst. This journal is © The Royal Society of Chemistry 2015 A differential protein solubility approach for the depletion of highly abundant proteins in plasma using ammonium sulfate Ravi Chand Bollineni1, 2*, Ingrid J. Guldvik3, Henrik Gronberg4, Fredrik Wiklund4, Ian G. Mills3, 5, 6 and Bernd Thiede1, 2 1Department of Biosciences, University of Oslo, Oslo, Norway 2Biotechnology Centre of Oslo, University of Oslo, Oslo, Norway 3Centre for Molecular Medicine Norway (NCMM), University of Oslo and Oslo University Hospitals, Norway 4Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm, Sweden 5Department of Cancer Prevention, Oslo University Hospitals, Oslo, Norway 6Department of Urology, Oslo University Hospitals, Oslo, Norway Keywords: ammonium sulfate, blood, depletion, plasma, protein precipitation *To whom the correspondence should be addressed: Ravi Chand Bollineni, Department of Biosciences, University of Oslo, P.O. Box 1066 Blindern, 0316 Oslo, Norway, Tel.: +47-22840512; Fax +47-22840501; E-mail: [email protected] Supplementary information Figure S1: SDS-PAGE analysis of serum proteins precipitated with the ethanol/sodium acetate (A), TCA/acetone (B) and ammonium sulfate precipitation (C). A) Ethanol/sodium acetate precipitation: (1) pellet obtained after 42% ethanol precipitation and (2) pellet obtained after precipitation of proteins in the supernatant with 0.8M sodium acetate (pH5.7) and (3) proteins left over in the supernatant. B) Serum proteins are precipitated with 10% TCA/acetone (1) and (2) proteins left over in the supernatant. C) Serum proteins are precipitated with increasing ammonium sulfate concentrations 15% (1), 25% (2), 35% (3), 40% (4), 45% (5), 50% (6) and total serum (7). -

Alantolactone Inhibits Cell Autophagy and Promotes Apoptosis Via AP2M1
Shi et al. Cancer Cell Int (2020) 20:442 https://doi.org/10.1186/s12935-020-01537-9 Cancer Cell International PRIMARY RESEARCH Open Access Alantolactone inhibits cell autophagy and promotes apoptosis via AP2M1 in acute lymphoblastic leukemia Ce Shi1†, Wenjia Lan1†, Zhenkun Wang1, Dongguang Yang1, Jia Wei1, Zhiyu Liu1, Yueqiu Teng1, Mengmeng Gu2, Tian Yuan3, Fenglin Cao1, Jin Zhou2 and Yang Li1* Abstract Background: Acute lymphoblastic leukemia (ALL) is an aggressive hematopoietic malignancy that is most com- monly observed in children. Alantolactone (ALT) has been reported to exhibit anti-tumor activity in diferent types of cancer. The aim of the present study was to investigate the anti-tumor activity and molecular mechanism of ALT in ALL. Methods: ALL cell lines were treated with 1, 5 and 10 μM ALT, and cell viability was assessed using an MTT assay and RNA sequencing. Flow cytometry, JC-1 staining and immunofuorescence staining assays were used to measure cell apoptosis and autophagy. Additionally, western blot analysis was used to detect expression of apoptosis and autophagy related proteins. Finally, the efects of ALT on tumor growth were assessed in a BV173 xenograft nude mouse model. Results: ALT inhibited the proliferation of ALL cells in a dose-dependent manner. Additionally, it was demonstrated that ALT inhibited cell proliferation, colony formation, autophagy, induced apoptosis and reduced tumor growth in vivo through upregulating the expression of adaptor related protein complex 2 subunit mu 1 (AP2M1). Moreover, the autophagy activator rapamycin, attenuated the pro-apoptotic efects of ALT on BV173 and NALM6 cell lines. Overexpression of AP2M1 decreased the expression of Beclin1 and the LC3-II/LC3-1 ratio, and increased p62 expres- sion. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Integrating Protein Copy Numbers with Interaction Networks to Quantify Stoichiometry in Mammalian Endocytosis
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Integrating protein copy numbers with interaction networks to quantify stoichiometry in mammalian endocytosis Daisy Duan1, Meretta Hanson1, David O. Holland2, Margaret E Johnson1* 1TC Jenkins Department of Biophysics, Johns Hopkins University, 3400 N Charles St, Baltimore, MD 21218. 2NIH, Bethesda, MD, 20892. *Corresponding Author: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Proteins that drive processes like clathrin-mediated endocytosis (CME) are expressed at various copy numbers within a cell, from hundreds (e.g. auxilin) to millions (e.g. clathrin). Between cell types with identical genomes, copy numbers further vary significantly both in absolute and relative abundance. These variations contain essential information about each protein’s function, but how significant are these variations and how can they be quantified to infer useful functional behavior? Here, we address this by quantifying the stoichiometry of proteins involved in the CME network. We find robust trends across three cell types in proteins that are sub- vs super-stoichiometric in terms of protein function, network topology (e.g. -
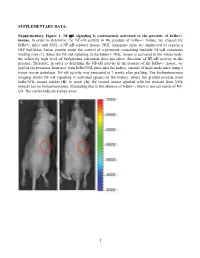
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

Β-Catenin-Mediated Wnt Signal Transduction Proceeds Through an Endocytosis-Independent Mechanism
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.13.948380; this version posted February 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. β-catenin-Mediated Wnt Signal Transduction Proceeds Through an Endocytosis-Independent Mechanism Ellen Youngsoo Rim1, , Leigh Katherine Kinney1, and Roel Nusse1, 1Howard Hughes Medical Institute, Department of Developmental Biology, Stanford University School of Medicine, Stanford, CA 94305, USA The Wnt pathway is a key intercellular signaling cascade that by GSK3β is inhibited. This leads to β-catenin accumulation regulates development, tissue homeostasis, and regeneration. in the cytoplasm and concomitant translocation into the nu- However, gaps remain in our understanding of the molecular cleus, where it can induce transcription of target genes. The events that take place between ligand-receptor binding and tar- importance of β-catenin stabilization in Wnt signal transduc- get gene transcription. Here we used a novel tool for quanti- tion has been demonstrated in many in vivo and in vitro con- tative, real-time assessment of endogenous pathway activation, texts (8, 9). However, immediate molecular responses to the measured in single cells, to answer an unresolved question in the ligand-receptor interaction and how they elicit accumulation field – whether receptor endocytosis is required for Wnt signal transduction. We combined knockdown or knockout of essential of β-catenin are not fully elucidated. components of Clathrin-mediated endocytosis with quantitative One point of uncertainty is whether receptor endocyto- assessment of Wnt signal transduction in mouse embryonic stem sis following Wnt binding is required for signal transduc- cells (mESCs). -

Carboxypeptidase B2 (CPB2) Human Shrna Lentiviral Particle (Locus ID 1361) Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TL305252V Carboxypeptidase B2 (CPB2) Human shRNA Lentiviral Particle (Locus ID 1361) Product data: Product Type: shRNA Lentiviral Particles Product Name: Carboxypeptidase B2 (CPB2) Human shRNA Lentiviral Particle (Locus ID 1361) Locus ID: 1361 Synonyms: CPU; PCPB; TAFI Vector: pGFP-C-shLenti (TR30023) Format: Lentiviral particles RefSeq: NM_001278541, NM_001872, NM_016413, NM_001872.1, NM_001872.2, NM_001872.3, NM_001872.4, NM_016413.1, NM_016413.2, NM_016413.3, NM_001278541.1, BC007057, BC007057.1, NM_001872.5 Summary: Carboxypeptidases are enzymes that hydrolyze C-terminal peptide bonds. The carboxypeptidase family includes metallo-, serine, and cysteine carboxypeptidases. According to their substrate specificity, these enzymes are referred to as carboxypeptidase A (cleaving aliphatic residues) or carboxypeptidase B (cleaving basic amino residues). The protein encoded by this gene is activated by trypsin and acts on carboxypeptidase B substrates. After thrombin activation, the mature protein downregulates fibrinolysis. Polymorphisms have been described for this gene and its promoter region. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jun 2013] shRNA Design: These shRNA constructs were designed against multiple splice variants at this gene locus. To be certain that your variant of interest is targeted, -

Host Cell Factors Necessary for Influenza a Infection: Meta-Analysis of Genome Wide Studies
Host Cell Factors Necessary for Influenza A Infection: Meta-Analysis of Genome Wide Studies Juliana S. Capitanio and Richard W. Wozniak Department of Cell Biology, Faculty of Medicine and Dentistry, University of Alberta Abstract: The Influenza A virus belongs to the Orthomyxoviridae family. Influenza virus infection occurs yearly in all countries of the world. It usually kills between 250,000 and 500,000 people and causes severe illness in millions more. Over the last century alone we have seen 3 global influenza pandemics. The great human and financial cost of this disease has made it the second most studied virus today, behind HIV. Recently, several genome-wide RNA interference studies have focused on identifying host molecules that participate in Influen- za infection. We used nine of these studies for this meta-analysis. Even though the overlap among genes identified in multiple screens was small, network analysis indicates that similar protein complexes and biological functions of the host were present. As a result, several host gene complexes important for the Influenza virus life cycle were identified. The biological function and the relevance of each identified protein complex in the Influenza virus life cycle is further detailed in this paper. Background and PA bound to the viral genome via nucleoprotein (NP). The viral core is enveloped by a lipid membrane derived from Influenza virus the host cell. The viral protein M1 underlies the membrane and anchors NEP/NS2. Hemagglutinin (HA), neuraminidase Viruses are the simplest life form on earth. They parasite host (NA), and M2 proteins are inserted into the envelope, facing organisms and subvert the host cellular machinery for differ- the viral exterior. -

Electronic Supplementary Material (ESI) for Analyst. This Journal Is © the Royal Society of Chemistry 2020
Electronic Supplementary Material (ESI) for Analyst. This journal is © The Royal Society of Chemistry 2020 Table S1. -

Functional Dependency Analysis Identifies Potential Druggable
cancers Article Functional Dependency Analysis Identifies Potential Druggable Targets in Acute Myeloid Leukemia 1, 1, 2 3 Yujia Zhou y , Gregory P. Takacs y , Jatinder K. Lamba , Christopher Vulpe and Christopher R. Cogle 1,* 1 Division of Hematology and Oncology, Department of Medicine, College of Medicine, University of Florida, Gainesville, FL 32610-0278, USA; yzhou1996@ufl.edu (Y.Z.); gtakacs@ufl.edu (G.P.T.) 2 Department of Pharmacotherapy and Translational Research, College of Pharmacy, University of Florida, Gainesville, FL 32610-0278, USA; [email protected]fl.edu 3 Department of Physiological Sciences, College of Veterinary Medicine, University of Florida, Gainesville, FL 32610-0278, USA; cvulpe@ufl.edu * Correspondence: [email protected]fl.edu; Tel.: +1-(352)-273-7493; Fax: +1-(352)-273-5006 Authors contributed equally. y Received: 3 November 2020; Accepted: 7 December 2020; Published: 10 December 2020 Simple Summary: New drugs are needed for treating acute myeloid leukemia (AML). We analyzed data from genome-edited leukemia cells to identify druggable targets. These targets were necessary for AML cell survival and had favorable binding sites for drug development. Two lists of genes are provided for target validation, drug discovery, and drug development. The deKO list contains gene-targets with existing compounds in development. The disKO list contains gene-targets without existing compounds yet and represent novel targets for drug discovery. Abstract: Refractory disease is a major challenge in treating patients with acute myeloid leukemia (AML). Whereas the armamentarium has expanded in the past few years for treating AML, long-term survival outcomes have yet to be proven. To further expand the arsenal for treating AML, we searched for druggable gene targets in AML by analyzing screening data from a lentiviral-based genome-wide pooled CRISPR-Cas9 library and gene knockout (KO) dependency scores in 15 AML cell lines (HEL, MV411, OCIAML2, THP1, NOMO1, EOL1, KASUMI1, NB4, OCIAML3, MOLM13, TF1, U937, F36P, AML193, P31FUJ). -

Common Gene Polymorphisms Associated with Thrombophilia
Chapter 5 Common Gene Polymorphisms Associated with Thrombophilia Christos Yapijakis, Zoe Serefoglou and Constantinos Voumvourakis Additional information is available at the end of the chapter http://dx.doi.org/10.5772/61859 Abstract Genetic association studies have revealed a correlation between DNA variations in genes encoding factors of the hemostatic system and thrombosis-related disease. Certain var‐ iant alleles of these genes that affect either gene expression or function of encoded protein are known to be genetic risk factors for thrombophilia. The chapter presents the current genetics and molecular biology knowledge of the most important DNA polymorphisms in thrombosis-related genes encoding coagulation factor V (FV), coagulation factor II (FII), coagulation factor XII (FXII), coagulation factor XIII A1 subunit (FXIIIA1), 5,10- methylene tetrahydrofolate reductase (MTHFR), serpine1 (SERPINE1), angiotensin I-con‐ verting enzyme (ACE), angiotensinogen (AGT), integrin A2 (ITGA2), plasma carboxypeptidase B2 (CPB2), platelet glycoprotein Ib α polypeptide (GP1BA), thrombo‐ modulin (THBD) and protein Z (PROZ). The molecular detection methods of each DNA polymorphism is presented, in addition to the current knowledge regarding its influence on thrombophilia and related thrombotic events, including stroke, myocardial infarction, deep vein thrombosis, spontaneous abortion, etc. In addition, best thrombosis prevention strategies with a combination of genetic counseling and molecular testing are discussed. Keywords: Thrombophilia, coagulation