In Situ Structural Studies of Tripeptidyl Peptidase II (TPPII) Reveal Spatial Association with Proteasomes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Catabolism Iii
Nitrogen Catabolism Glycogenolysis Protein Fat catabolism Catabolism Fatty Acid Amino-acid GlycolysisDegradation catabolism Pyruvate Oxidation Krebs' Cycle Phosphorylation Oxidative CATABOLISM III: Digestion and Utilization of Proteins • Protein degradation • Protein turnover – The ubiquitin pathway – Protein turnover is tightly regulated • Elimination of nitrogen – By fish, flesh and fowl – How is the N of amino acids liberated and eliminated? • How are amino acids oxidized for energy 1 Protein Catabolism Sources of AMINO ACIDS: •Dietary amino acids that exceed body’s protein synthesis needs •Excess amino acids from protein turnover (e.g., proteolysis and regeneration of proteins) •Proteins in the body can be broken down (muscle wasting) to supply amino acids for energy when carbohydrates are scarce (starvation, diabetes mellitus). •Carnivores use amino acids for energy more than herbivores, plants, and most microorganisms Protein Catabolism The Digestion Pathway • Pro-enzymes are secreted (zymogens) and the environment activates them by specific proteolysis. • Pepsin hydrolyzes protein into peptides in the stomach. • Trypsin and chymotrypsin hydrolyze proteins and larger peptides into smaller peptides in the small intestine. • Aminopeptidase and carboxypeptidases A and B degrade peptides into amino acids in the small intestine. 2 Protein Catabolism The Lysosomal Pathway • Endocytosis, either receptor-mediated, phagocytosis, or pinocytosis engulfs extra- cellular proteins into vesicles. • These internal vesicles fuse as an early endosome. • This early endosome is acidified by the KFERQ Substrates vATPase (“v” for vesicular). • Components that are recycled, like receptors, HSPA8 are sequestered in smaller vesicles to create Co-chaperones the multivesicular body (MVB), sometimes called a late endosome. • If set for degradation, it will fuse with a KFERQ primary lysosome (red) which contains many cathepsin-type proteases. -

JASON MARC GOLDSTEIN the Isolation, Characterization
JASON MARC GOLDSTEIN The Isolation, Characterization and Cloning of Three Novel Peptidases From Streptoccocus gordonii: Their Potential Roles in Subacute Bacterial Endocarditis (Under the Direction of JAMES TRAVIS) Streptococcus gordonii is generally considered a benign inhabitant of the oral microflora yet is a primary etiological agent in the development of subacute bacterial endocarditis (SBE), an inflammatory state that propagates thrombus formation and tissue damage on the surface of heart valves. Colonization and adherence mechanisms have been identified, yet factors necessary to sustain growth remain unidentified. Strain FSS2 produced three extracellular aminopeptidase activities during growth in neutral pH- controlled batch cultures. The first included a serine-class dipeptidyl-aminopeptidase, an x-Pro DPP (Sg-xPDPP) found as an 85 kDa monomer by SDS-PAGE while appearing as a homodimer under native conditions. Kinetic studies indicated a unique and stringent x- Pro specificity comparable to the DPPIV/CD26 and lactococcal x-Pro DPP families. Isolation of the full-length gene uncovered a 759-amino acid polypeptide with a mass of 87,115 Da and theoretical pI of 5.6. Significant homology was found with PepX gene family members from Lactobacillus ssp. and Lactococcus ssp., and putative streptococcal x-Pro DPPs. The second activity was a putative serine-class arginine aminopeptidase (Sg- RAP) with some cysteine-class characteristics. It was found as a protein monomer of 70 kDa under denaturing conditions. Nested PCR cloning enabled the isolation of a 324 bp- long DNA fragment encoding the protein’s 108 amino acid N-terminus. Culture activity profiles and N-terminal sequence analysis indicated the release of this protein from the cell surface. -

Effect of Dietary Protein on Lipid and Glucose Metabolism: Implications for Metabolic Health
Effect of dietary protein on lipid and glucose metabolism: implications for metabolic health Annemarie Rietman 34334 Rietman.indd 1 09-07-15 09:32 Thesis committee Promotors Prof. Dr F.J. Kok Professor of Nutrition and Health Wageningen University Prof. Dr D. Tomé Professor of Nutrition and Protein Metabolism AgroParisTech, France Co-promotor Dr M. Mensink Assistant professor, Division of Human Nutrition Other members Prof. Dr W.H. Hendriks, Wageningen University Prof. Dr E. Blaak, Maastricht University, The Netherlands Prof. L. Tappy, Université de Lausanne, Switzerland Dr R. Heiligenberg, Hospital ‘De Gelderse Vallei’, Ede, The Netherlands This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). 34334 Rietman.indd 2 09-07-15 09:32 Effect of dietary protein on lipid and glucose metabolism: implications for metabolic health Annemarie Rietman Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr A.P.J. Mol, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 11 September 2015 at 4 p.m. in the Aula. 34334 Rietman.indd 3 09-07-15 09:32 Annemarie Rietman Effect of dietary protein on lipid and glucose metabolism: implications for metabolic health 160 pages PhD thesis, Wageningen University, Wageningen, NL (2015) With references, with summaries in English and Dutch ISBN 978-94-6257-348-2 34334 Rietman.indd 4 09-07-15 11:15 Abstract Background: Diet is an important factor in the development of the Metabolic Syndrome (Mets) and type 2 Diabetes Mellitus. -

High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma
molecules Article High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma Elisa Maffioli 1,2 , Zhenze Jiang 3, Simona Nonnis 1,2 , Armando Negri 1,2, Valentina Romeo 1, Christopher B. Lietz 3, Vivian Hook 3,4, Giuseppe Ristagno 5, Giuseppe Baselli 6, Erik B. Kistler 7,8 , Federico Aletti 9, Anthony J. O’Donoghue 3,* and Gabriella Tedeschi 1,2,* 1 Department of Veterinary Medicine, University of Milano, 20133 Milano, Italy; elisa.maffi[email protected] (E.M.); [email protected] (S.N.); [email protected] (A.N.); [email protected] (V.R.) 2 Centre for Nanostructured Materials and Interfaces (CIMAINA), University of Milano, 20133 Milano, Italy 3 Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92093, USA; [email protected] (Z.J.); [email protected] (C.B.L.); [email protected] (V.H.) 4 Department of Neurosciences, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA 5 Department of Pathophysiology and Transplantation, University of Milan, 20133 Milan, Italy; [email protected] 6 Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, 20133 Milan, Italy; [email protected] 7 Department of Anesthesiology & Critical Care, University of California San Diego, La Jolla, CA 92093, USA; [email protected] 8 Department of Anesthesiology & Critical Care, VA San Diego HealthCare System, San Diego, CA 92161, USA 9 Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA; [email protected] * Correspondence: [email protected] (A.J.O.); [email protected] (G.T.); Tel.: +1-8585345360 (A.J.O.); +39-02-50318127 (G.T.) Academic Editor: Paolo Iadarola Received: 28 July 2020; Accepted: 4 September 2020; Published: 6 September 2020 Abstract: Proteomic technologies have identified 234 peptidases in plasma but little quantitative information about the proteolytic activity has been uncovered. -

Gluconeogenesis 1
GLUCONEOGENESIS It is the process by which glucose molecules are produced from non- carbohydrate precursors, which include lactate, glucogenic amino acids, glycerol part of fat and propionyl CoA derived from odd chain fatty acids. Gluconeogenesis occurs mainly in the liver, and to a lesser extent in the kidney. The pathway is partly mitochondrial and partly cytoplasmic. 4 3 2 ! 1 Gluconeogenesis involves several enzymes of glycolysis, but it is not a reversal of glycolysis. The irreversible steps in glycolysis are circumvented by four enzymes which are designated as the key enzymes of gluconeogenesis, these enzymes are: 1. Pyruvate Carboxylase 1 http://cden.tu.edu.iq Tikrit University College of Dentistry Pyruvate in the cytoplasm enters the mitochondria. Then, carboxylation of pyruvate to oxaloacetate is catalysed by a mitochondrial enzyme, Pyruvate carboxylase. It needs the co-enzymes biotin and ATP. 2. Phosphoenol Pyruvate Carboxy Kinase (PEPCK) In the cytoplasm, PEPCK enzyme converts oxaloacetate to phosphoenol pyruvate by removing a molecule of CO2. GTP donates the phosphate. The net effect of these two reactions is the conversion of pyruvate to phosphoenol pyruvate. This circumvents the irreversible step 9 of glycolysis. Partial Reversal of Glycolysis The phosphoenol pyruvate undergoes further reactions catalyzed by the glycolytic enzymes to form fructose-1,6-bisphosphate (see glycolysis steps 8,7,6,5 and 4; all these reactions are freely reversible). 3. Fructose-1, 6-bisphosphatase Fructose-1, 6-bisphosphate is then converted by fructose 1,6- bisphosphatase to form fructose -6-phosphate. This will bypass the step of PFK reaction (see step 3 of glycolysis). Then fructose-6-phosphate is isomerized to glucose-6-phosphate by the freely reversible reaction catalyzed by hexose phosphate isomerase (step2 in glycolysis). -

Lipid Metabolism - 1
Lipid metabolism - 1. Catabolism of triacylglycerols, oxidation of fatty acids and glycerol. Ketogenesis. ASSOC. PROF. BILETS M.V. Lecture plan Catabolism of triacylglycerols in adipocytes of adipose tissue. Biosynthesis of triacylglycerols. Oxidation of glycerol: enzyme reactions, bioenergetics. Lipoproteins. Hyperlipoproteinemias. Oxidation of glycerol: enzyme reactions, bioenergetics. Oxidation of fatty acids (β-oxidation). Ketone bodies. Biosynthesis of fatty acids. Lipids are organic compounds, are nonpolar molecules, which are soluble only in nonpolar solvents and insoluble in water. Types of Lipids: Simple Lipids Esters of fatty acids with various alcohols. Fats: Esters of fatty acids with glycerol. Oils are fats in the liquid state Steroids (cholesterol) Complex Lipids Esters of fatty acids containing groups in addition to alcohol and a fatty acid. Phospholipids: These are lipids containing, in addition to fatty acids and alcohol, a phosphoric acid residue. They frequently have nitrogen-containing bases and other substituents, eg, in glycerophospholipids the alcohol is glycerol and in sphingophospholipids the alcohol is sphingosine. Glycolipids (glycosphingolipids): Lipids containing a fatty acid, sphingosine, and carbohydrate. Structure of triacylglycerol https://www.researchgate.net/figure/Triacylglycerol-TAG-structure-showing-glycerol-with- three-fatty-acids_fig1_316787887 Lipolysis is the metabolic pathway through which lipid triacylglycerols are hydrolyzed into a glycerol and three fatty acids. Occurs in adipocytes. Lipolysis -
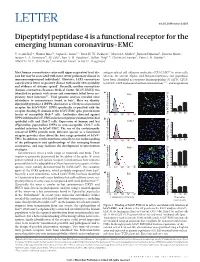
Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC
LETTER doi:10.1038/nature12005 Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC V. Stalin Raj1*, Huihui Mou2*, Saskia L. Smits1,3, Dick H. W. Dekkers4, Marcel A. Mu¨ller5, Ronald Dijkman6, Doreen Muth5, Jeroen A. A. Demmers4, Ali Zaki7, Ron A. M. Fouchier1, Volker Thiel6,8, Christian Drosten5, Peter J. M. Rottier2, Albert D. M. E. Osterhaus1, Berend Jan Bosch2 & Bart L. Haagmans1 Most human coronaviruses cause mild upper respiratory tract dis- antigen-related cell adhesion molecules (CEACAM)10 to enter cells, ease but may be associated with more severe pulmonary disease in whereas for several Alpha- and Betacoronaviruses, two peptidases immunocompromised individuals1. However, SARS coronavirus have been identified as receptors (aminopeptidase N (APN, CD13) caused severe lower respiratory disease with nearly 10% mortality for hCoV-229E and several animal coronaviruses11,12, and angiotensin and evidence of systemic spread2. Recently, another coronavirus (human coronavirus-Erasmus Medical Center (hCoV-EMC)) was a identified in patients with severe and sometimes lethal lower res- )] Vero –1 piratory tract infection3,4. Viral genome analysis revealed close 9 ml 8 5 50 relatedness to coronaviruses found in bats . Here we identify 7 dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional 6 5 receptor for hCoV-EMC. DPP4 specifically co-purified with the 4 receptor-binding S1 domain of the hCoV-EMC spike protein from 3 Relative cell number log[GE (TCID 0 20 40 lysates of susceptible Huh-7 cells. Antibodies directed against 0 101 102 103 104 DPP4 inhibited hCoV-EMC infection of primary human bronchial b )] COS-7 –1 epithelial cells and Huh-7 cells. -

And Exopeptidases in a Processing Enzyme System: Activation
Proc. Nail. Acad. Sci. USA Vol. 85, pp. 5468-5472, August 1988 Biochemistry Relationship between endo- and exopeptidases in a processing enzyme system: Activation of an endoprotease by the aminopeptidase B-like activity in somatostatin-28 convertase (brain cortex/basic amino acid pairs/peptide substrates/protease inhibitors/prohormone maturation) SOPHIE GOMEZ, PABLO GLUSCHANKOF, AGNES LEPAGE, AND PAUL COHEN Groupe de Neurobiochimie Cellulaire et Moldculaire, Universitd Pierre et Marie Curie, Unitd Associde 554 au Centre National de la Recherche Scientifique, 96 boulevard Raspail, 75006 Paris, France Communicated by I. Robert Lehman, April 8, 1988 (receivedfor review December 15, 1987) ABSTRACT The somatostatin-28 convertase activity in- somatostatin-14 and the amino-terminal dodecapeptide so- volved in vitro in the processing of somatostatin-28 into the matostatin-28-(1-12) (16). neuropeptides somatostatin-28-(1-12) and somatostatin-14 is We have described an endoprotease that cleaves the composed of an endoprotease and a basic aminopeptidase. We peptide bond on the amino side ofthe Arg-Lys doublet in the report herein on the purification to apparent homogeneity of somatostatin-28 sequence (17, 18), releasing the somato- these two constituents and on their functional interrelationship. statin-28-(1-12) fragment and [Arg-2,Lys-1]somatostatin-14. In particular we observed that after various physicochemical The released [Arg-2,Lys-']somatostatin-14 is further proc- treatments, the 90-kDa endoprotease activity was recovered essed by an aminopeptidase B-like activity (18, 19) that is also both at this molecular mass and as a 45-kDa entity. Moreover, present in the preparation. We report herein the purification the production of [Arg2,LysJllsomatostatin-14 from somato- to apparent homogeneity ofthese two activities. -

The Postabsorptive State
Chapter 26 Lecture Outline See separate PowerPoint slides for all figures and tables pre- inserted into PowerPoint without notes. Copyright © McGraw-Hill Education. Permission required for reproduction or display. 1 Introduction • Nutrition is the starting point and the basis for all human form and function – The source of fuel that provides energy for all biological work – The source of raw materials for replacement of worn- out biomolecules and cells • Metabolism is the chemical change that lies at the foundation of form and function 26-2 Nutrition • Expected Learning Outcomes – Describe some factors that regulate hunger and satiety. – Define nutrient and list the six major categories of nutrients. – State the function of each class of macronutrients, the approximate amounts required in the diet, and some major dietary sources of each. – Name the blood lipoproteins, state their functions, and describe how they differ from each other. – Name the major vitamins and minerals required by the body and the general functions they serve. 26-3 Body Weight and Energy Balance • Weight—determined by the body’s energy balance – If energy intake and output are equal, body weight is stable – Gain weight if intake exceeds output – Lose weight if output exceeds intake – Weight seems to have a stable, homeostatic set point • Varies from person to person • Combination of heredity and environmental influences – 30% to 50% of variation in human weight is hereditary – Environmental factors such as eating and exercise habits account for the rest of the variation -

Proteolytic Cleavage—Mechanisms, Function
Review Cite This: Chem. Rev. 2018, 118, 1137−1168 pubs.acs.org/CR Proteolytic CleavageMechanisms, Function, and “Omic” Approaches for a Near-Ubiquitous Posttranslational Modification Theo Klein,†,⊥ Ulrich Eckhard,†,§ Antoine Dufour,†,¶ Nestor Solis,† and Christopher M. Overall*,†,‡ † ‡ Life Sciences Institute, Department of Oral Biological and Medical Sciences, and Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, British Columbia V6T 1Z4, Canada ABSTRACT: Proteases enzymatically hydrolyze peptide bonds in substrate proteins, resulting in a widespread, irreversible posttranslational modification of the protein’s structure and biological function. Often regarded as a mere degradative mechanism in destruction of proteins or turnover in maintaining physiological homeostasis, recent research in the field of degradomics has led to the recognition of two main yet unexpected concepts. First, that targeted, limited proteolytic cleavage events by a wide repertoire of proteases are pivotal regulators of most, if not all, physiological and pathological processes. Second, an unexpected in vivo abundance of stable cleaved proteins revealed pervasive, functionally relevant protein processing in normal and diseased tissuefrom 40 to 70% of proteins also occur in vivo as distinct stable proteoforms with undocumented N- or C- termini, meaning these proteoforms are stable functional cleavage products, most with unknown functional implications. In this Review, we discuss the structural biology aspects and mechanisms -
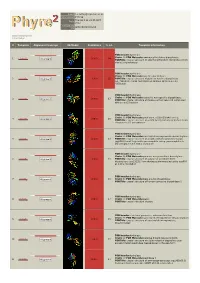
Phyre 2 Results for P15288
Email [email protected] Description P15288 Thu Jan 5 11:34:45 GMT Date 2012 Unique Job d2087d8d303c52c9 ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:hydrolase Chain: B: PDB Molecule:aminoacyl-histidine dipeptidase; 1 c3mruB_ 100.0 64 Alignment PDBTitle: crystal structure of aminoacylhistidine dipeptidase from vibrio2 alginolyticus PDB header:hydrolase Chain: B: PDB Molecule:xaa-his dipeptidase; 2 c2qyvB_ Alignment 100.0 55 PDBTitle: crystal structure of putative xaa-his dipeptidase (yp_718209.1) from2 haemophilus somnus 129pt at 2.11 a resolution PDB header:hydrolase Chain: A: PDB Molecule:cytosolic non-specific dipeptidase; 3 c2zogA_ 100.0 17 Alignment PDBTitle: crystal structure of mouse carnosinase cn2 complexed with zn and2 bestatin PDB header:hydrolase Chain: B: PDB Molecule:peptidase, m20/m25/m40 family; 4 c2pokB_ 100.0 16 Alignment PDBTitle: crystal structure of a m20 family metallo peptidase from streptococcus2 pneumoniae PDB header:hydrolase Chain: A: PDB Molecule:succinyl-diaminopimelate desuccinylase; 5 c3pfeA_ Alignment 100.0 13 PDBTitle: crystal structure of a m20a metallo peptidase (dape, lpg0809) from2 legionella pneumophila subsp. pneumophila str. philadelphia 1 at 1.503 a resolution PDB header:hydrolase Chain: B: PDB Molecule:putative acetylornithine deacetylase; 6 c3pfoB_ Alignment 100.0 16 PDBTitle: crystal structure of a putative acetylornithine deacetylase (rpa2325)2 from rhodopseudomonas palustris cga009 at 1.90 a resolution PDB -

Branched Chain Amino Acid Catabolism Fuels Adipocyte Differentiation and Lipogenesis
CORE Metadata, citation and similar papers at core.ac.uk Provided by eScholarship - University of California UCLA UCLA Previously Published Works Title Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Permalink https://escholarship.org/uc/item/6zd459ds Journal Nature chemical biology, 12(1) ISSN 1552-4450 Authors Green, Courtney R Wallace, Martina Divakaruni, Ajit S et al. Publication Date 2016 DOI 10.1038/nchembio.1961 Peer reviewed eScholarship.org Powered by the California Digital Library University of California HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Nat Chem Manuscript Author Biol. Author Manuscript Author manuscript; available in PMC 2016 May 18. Published in final edited form as: Nat Chem Biol. 2016 January ; 12(1): 15–21. doi:10.1038/nchembio.1961. Branched chain amino acid catabolism fuels adipocyte differentiation and lipogenesis Courtney R. Greena, Martina Wallacea, Ajit S. Divakarunib, Susan A. Phillipsc,d, Anne N. Murphyb, Theodore P. Ciaraldic,d, and Christian M. Metallo*,a,e aDepartment of Bioengineering, University of California–San Diego, La Jolla, CA 92093 bDepartment of Pharmacology, University of California–San Diego, La Jolla, CA 92093 cVeterans Affairs San Diego Healthcare System, San Diego, CA; Department of Medicine, University of California, San Diego, La Jolla, CA dMedicine, University of California, San Diego, La Jolla, CA eInstitute of Engineering in Medicine, University of California, San Diego, La Jolla, CA Abstract Adipose tissue plays important roles in regulating carbohydrate and lipid homeostasis, though less is known about the regulation of amino acid metabolism in adipocytes. Here we applied isotope tracing to pre–adipocytes and differentiated adipocytes to quantify the contributions of different substrates to tricarboxylic acid metabolism and lipogenesis.