Pathways, Mechanisms, and Rates of Polyploid Formation in Flowering Plants
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Second Contribution to the Vascular Flora of the Sevastopol Area
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Wulfenia Jahr/Year: 2015 Band/Volume: 22 Autor(en)/Author(s): Seregin Alexey P., Yevseyenkow Pavel E., Svirin Sergey A., Fateryga Alexander Artikel/Article: Second contribution to the vascular flora of the Sevastopol area (the Crimea) 33-82 © Landesmuseum für Kärnten; download www.landesmuseum.ktn.gv.at/wulfenia; www.zobodat.at Wulfenia 22 (2015): 33 – 82 Mitteilungen des Kärntner Botanikzentrums Klagenfurt Second contribution to the vascular flora of the Sevastopol area (the Crimea) Alexey P. Seregin, Pavel E. Yevseyenkov, Sergey A. Svirin & Alexander V. Fateryga Summary: We report 323 new vascular plant species for the Sevastopol area, an administrative unit in the south-western Crimea. Records of 204 species are confirmed by herbarium specimens, 60 species have been reported recently in literature and 59 species have been either photographed or recorded in field in 2008 –2014. Seventeen species and nothospecies are new records for the Crimea: Bupleurum veronense, Lemna turionifera, Typha austro-orientalis, Tyrimnus leucographus, × Agrotrigia hajastanica, Arctium × ambiguum, A. × mixtum, Potamogeton × angustifolius, P. × salicifolius (natives and archaeophytes); Bupleurum baldense, Campsis radicans, Clematis orientalis, Corispermum hyssopifolium, Halimodendron halodendron, Sagina apetala, Solidago gigantea, Ulmus pumila (aliens). Recently discovered Calystegia soldanella which was considered to be extinct in the Crimea is the most important confirmation of historical records. The Sevastopol area is one of the most floristically diverse areas of Eastern Europe with 1859 currently known species. Keywords: Crimea, checklist, local flora, taxonomy, new records A checklist of vascular plants recorded in the Sevastopol area was published seven years ago (Seregin 2008). -

Динамика Демографической Структуры Ценопопуляций Tulipa Suaveolens Roth (Liliaceae, Magnoliophyta) В Нижнем Поволжье
ПОВОЛЖСКИЙ ЭКОЛОГИЧЕСКИЙ ЖУРНАЛ. 2019. № 3. С. 291 – 310 УДК 581.8 ДИНАМИКА ДЕМОГРАФИЧЕСКОЙ СТРУКТУРЫ ЦЕНОПОПУЛЯЦИЙ TULIPA SUAVEOLENS ROTH (LILIACEAE, MAGNOLIOPHYTA) В НИЖНЕМ ПОВОЛЖЬЕ А. С. Кашин, Н. А. Петрова, И. В. Шилова, А. С. Пархоменко Ботанический сад Саратовского национального исследовательского государственного университета имени Н. Г. Чернышевского Россия, 410010, Саратов, Навашина E-mail: [email protected] Поступила в редакцию 15.09.2018 г., после доработки 14.12.2018 г., принята 15.02.2019 г. Кашин А. С., Петрова Н. А., Шилова И. В., Пархоменко А. С. Динамика демографической структуры ценопопуляций Tulipa suaveolens Roth (Liliaceae, Magnoliophyta) в Нижнем По- волжье // Поволжский экологический журнал. 2019. № 3. С. 291 – 310. DOI: https://doi.org/10.35885/1684-7318-2019-3-291-310 Исследована демографическая структура 39 ценопопуляций Tulipa suaveolens Roth в Нижнем Поволжье. Показано, что они занимают площадь от 0.01 до 20000 и более га. При этом малые по площади популяции произрастают в основном ближе к северной границе ареала вида. Плотность всех особей (1.6 – 240.7 экз./м2) и количество генеративных расте- ний (0.1 – 58.2 экз./м2) на межпопуляционном уровне варьировали в широком диапазоне, но по годам существенно изменялись преимущественно в популяциях, подверженных рекреа- ционной нагрузке или выпасу. Доля генеративных особей составляла от 2 до 96%. При этом в 2013 – 2016 гг. имела место значимая отрицательная корреляция между географической широтой, соответствующей месту нахождения ценопопуляций, и долей растений генера- тивного состояния в них. Напротив, в 2017 – 2018 гг. на юге и западе исследуемой части ареала преобладали растения прегенеративного периода и существенно снизилась доля цве- тущих растений. Наблюдавшаяся динамика хорошо соотносится с погодными условиями периодов вегетации тюльпанов. -

Reproduction and Identification of Root-Knot Nematodes on Perennial Ornamental Plants in Florida
REPRODUCTION AND IDENTIFICATION OF ROOT-KNOT NEMATODES ON PERENNIAL ORNAMENTAL PLANTS IN FLORIDA By ROI LEVIN A THESIS PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE UNIVERSITY OF FLORIDA 2005 Copyright 2005 by Roi Levin ACKNOWLEDGMENTS I would like to thank my chair, Dr. W. T. Crow, and my committee members, Dr. J. A. Brito, Dr. R. K. Schoellhorn, and Dr. A. F. Wysocki, for their guidance and support of this work. I am honored to have worked under their supervision and commend them for their efforts and contributions to their respective fields. I would also like to thank my parents. Through my childhood and adult years, they have continuously encouraged me to pursue my interests and dreams, and, under their guidance, gave me the freedom to steer opportunities, curiosities, and decisions as I saw fit. Most of all, I would like to thank my fiancée, Melissa A. Weichert. Over the past few years, she has supported, encouraged, and loved me, through good times and bad. I will always remember her dedication, patience, and sacrifice while I was working on this study. I would not be the person I am today without our relationship and love. iii TABLE OF CONTENTS page ACKNOWLEDGMENTS ................................................................................................. iii LIST OF TABLES............................................................................................................. vi LIST OF FIGURES .......................................................................................................... -
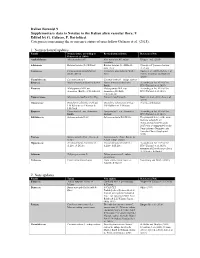
9 Edited by G. Galasso, F. Bartolucci Categories Concerning the Occurrence Status of Taxa Follow Galasso Et Al
Italian Botanist 9 Supplementary data to Notulae to the Italian alien vascular flora: 9 Edited by G. Galasso, F. Bartolucci Categories concerning the occurrence status of taxa follow Galasso et al. (2018). 1. Nomenclatural updates Family Nomenclature according to Revised nomenclature References/Note Galasso et al. (2018) Asphodelaceae Aloë maculata All. Aloë maculata All. subsp. Klopper et al. (2020) maculata Solanaceae Batatas batatas (L.) H.Karst. Batatas batatas (L.) H.Karst., Synonym of Ipomoea batatas nom. inval. (L.) Lam. Lauraceae Cinnamomum glanduliferum Camphora glandulifera (Wall.) Huang et al. (2016), Rohde et al. (Wall.) Meisn. Nees (2017), Trofimov and Rohwer (2020) Cucurbitaceae Cucumis sativus L. Cucumis sativus L. subsp. sativus Rosaceae Malus domestica (Borkh.) Borkh. Malus domestica (Suckow) According to Art. 41.4 of the Borkh. ICN (Turland et al. 2018) Rosaceae Malus pumila Mill. var. Malus pumila Mill. var. According to Art. 41.4 of the domestica (Borkh.) C.K.Schneid. domestica (Suckow) ICN (Turland et al. 2018) C.K.Schneid. Papaveraceae Meconopsis cambrica (L.) Vig. Papaver cambricum L. Kadereit et al. (2011), Liu et al. (2014) Onagraceae Oenothera oakesiana (A.Gray) Oenothera oakesiana (A.Gray) Priority combination J.W.Robbins ex S.Watson & J.W.Robbins ex S.Watson J.M.Coult. Rosaceae Pyrus malus L. var. domestica Pyrus malus L. var. domestica According to Art. 41.4 of the Borkh. Suckow ICN (Turland et al. 2018) Salviniaceae Salvinia adnata Desv. Salvinia molesta D.S.Mitch. The proposal to reject the name Salvinia adnata Desv. (Schwartsburd and Miranda 2017) was recommended by the Nomenclature Committee for Vascular Plants (Applequist 2019) Poaceae Setaria pumila (Poir.) Roem. -
ISTA List of Stabilized Plant Names 6Th Edition
ISTA List of Stabilized Plant Names 6th Edition ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema Published by All rights reserved. No part of this publication may The International Seed Testing Association (ISTA) be reproduced, stored in any retrieval system or Zürichstr. 50, CH-8303 Bassersdorf, Switzerland transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, ©2014 International Seed Testing Association (ISTA) without prior permission in writing from ISTA. ISBN 978-3-906549-77-4 ISTA List of Stabilized Plant Names 1st Edition 1966 ISTA Nomenclature Committee Chair: Prof. P. A. Linehan 2nd Edition 1983 ISTA Nomenclature Committee Chair: Dr. H. Pirson 3rd Edition 1988 ISTA Nomenclature Committee Chair: Dr. W. A. Brandenburg 4th Edition 2001 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 5th Edition 2007 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 6th Edition 2013 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema ii 6th Edition 2013 ISTA List of Stabilized Plant Names Contents Contents Preface ...................................................... iv L ................................................................41 Acknowledgements .................................... v M ...............................................................46 Symbols and abbreviations ....................... vi N ...............................................................50 ISTA List of Stabilized Plant Names ........... 1 O ...............................................................51 -

Flora of Russia" on Inaturalist: a Dataset
PREPRINT Posted on 02/10/2020 DOI: https://doi.org/10.3897/arphapreprints.e59250 "Flora of Russia" on iNaturalist: a dataset Alexey Seregin, Dmitriy Bochkov, Julia Shner, Eduard Garin, Igor Pospelov, Vadim Prokhorov, Pavel Golyakov, Sergey Mayorov, Sergey Svirin, Alexander Khimin, Marina Gorbunova, Ekaterina Kashirina, Olga Kuryakova, Boris Bolshakov, Aleksandr Ebel, Anatoliy Khapugin, Maxim Mallaliev, Sergey Mirvoda, Sergey Lednev, Dina Nesterkova, Nadezhda Zelenova, Svetlana Nesterova, Viktoriya Zelenkova, Georgy Vinogradov, Olga Biryukova, Alla Verkhozina, Alexey Zyrianov, Sergey Gerasimov, Ramazan Murtazaliev, Yurii Basov, Kira Marchenkova, Dmitry Vladimirov, Dina Safina, Sergey Dudov, Nikolai Degtyarev, Diana Tretyakova, Daba Chimitov, Evgenij Sklyar, Alesya Kandaurova, Svetlana Bogdanovich, Alexander Dubynin, Olga Chernyagina, Aleksandr Lebedev, Mikhail Knyazev, Irina Mitjushina, Nina Filippova, Kseniia Dudova, Igor Kuzmin, Tatyana Svetasheva, Vladimir Zakharov, Vladimir Travkin, Yaroslav Magazov, Vladimir Teploukhov, Andrey Efremov, Olesya Deineko, Viktor Stepanov, Eugene Popov, Dmitry Kuzmenckin, Tatiana Strus, Tatyana Zarubo, Konstantin Romanov, Alexei Ebel, Denis Tishin, Vladimir Arkhipov, Vladimir Korotkov, Svetlana Kutueva, Vladimir Gostev, Mikhail Krivosheev, Natalia Gamova, Veronica Belova, Oleg Kosterin, Sergey Prokopenko, Rinat Sultanov, Irina Kobuzeva, Nikolay Dorofeev, Alexander Yakovlev, Yuriy Danilevsky, Irina Zolotukhina, Damir Yumagulov, Valerii Glazunov, Vladimir Bakutov, Andrey Danilin, Igor Pavlov, Elena Pushay, -

Skvortsovia: 6(2): 1–66 (2020) Skvortsovia ISSN 2309-6497 (Print) Copyright: © 2020 Russian Academy of Sciences ISSN 2309-6500 (Online)
Skvortsovia: 6(2): 1–66 (2020) Skvortsovia ISSN 2309-6497 (Print) Copyright: © 2020 Russian Academy of Sciences http://skvortsovia.uran.ru/ ISSN 2309-6500 (Online) Conference Proceedings The Ninth Conference in Memory of Alexei K. Skvortsov Tsitsin Main Botanical Garden, Russian Academy of Sciences, Moscow. February, 2020 Translation received: 22 August 2020 | Accepted by Ivan A. Schanzer: 26 August 2020 | Published online: 4 September 2020 Translated by: Irina Kadis. Edited by: Irina Belyaeva and Keith Chamberlain Conference Overview Ivan A. Schanzer Tsitsin Main Botanical Garden of Russian Academy of Sciences, Botanicheskaya St. 4, 127276 Moscow, Russian Federation Email: [email protected] The 9th annual Conference to celebrate the Centenary of Professor Alexey K. Skvortsov (1920–2008) was held at Tsitsin Main Botanical Garden of the Russian Academy of Sciences (MBG) in February 2020. The Conference was organized by the Moscow Branch of the Russian Botanical Society and supported by Tsitsin Main Botanical Garden and Moscow State University (MSU). Financial support was provided by the Chimmed Group, Moscow, and the Russian Foundation for Basic Research (grant No 20-04-20001). Unlike the previous conferences, the topic was widened to encompass several of the areas of interest to Prof. Skvortsov, namely, microevolution, taxonomy, and flora studies. The Conference took the form of a two-day meeting with an additional short lecture course on plant nomenclature given by Irina Belyaeva Royal Botanic Gardens, Kew (RBG, Kew). The 9th Conference gathered botanists from the Russian regions of Moscow, St. Petersburg, Arzamas, Balashov, Borok, Bryansk, Kalach-na-Donu, Magadan, Novosibirsk, Penza, Ryazan, Saratov, Ufa, and Volgograd, as well as Debrecen (Hungary), London (UK), Storrs (Connecticut, US), and Vilnius (Lithuania), 87 in all, who presented their talks and posters, and took part in discussions. -

Scarlet (Tulipa Suaveolensroth). by WILL EM EDUARD DE MOL (Amsterdam)
Pro ducing at will of fertile diploid and tetraploid gametes in Duc van Thol, Scarlet (Tulipa suaveolensRoth). by WILL EM EDUARD DE MOL (Amsterdam). With 22 figures in text and on plates IV and V. Manuskript eingegangen am 9. Dezember 1927. I. Introduction. II. Duc van Thol, Scarlet and its origin. III. The karyologic research of the pollen-grains. IV. A constant relation between the number of chromosomes and the surface of the nucleus and that between the number of chromosomes and the surface of the tell. V. The germinating of the monoploid, diploid and tetraploid pollen-grains. VI. The isolation of the diploid and tetraploid pollen-grains. VII. The pollination-experiments and their result. VIII. Perspectives. IX. Summary in German (Zusammenfassung). References. Explanation of figures. I. Introduction. It is a great pleasure to the writer to have this paper published in the Vierteljahrsschrift der Naturforschenden Ge- sellschaft in Zürich, on the Occasion of the 70th anniversary of his esteemed teacher and friend Professor HANS SCHINZ. In 1920 and 1921 through particular external causes the experiment succeeded to cause in the Hyacinth the development of pollen-grains with a larger number of nuclei than normal, or with nuclei which contained the somatic number of chromo- somes instead of the haploid number. Later, in 1923, the attempt to originate triploid off spring from diploid parents by pollination with duplicated pollen-grains was crowned with success. See nos. 1921-1926 a, 1927 c-1927 e and 1928 b. 74 Fenschrift HANS SCHINZ. In 1923 similar experiments were begun at the Tulips. The results hereof have partially been published in short notes and papers. -

297313 VOL2.Pdf
THE DEVELOPMENT OF HERBACEOUS PLANTING IN BRITAIN AND GERMANY FROM THE NINETEENTH TO EARLY TWENTIETH CENTURY Volume 11 Of 2 Volumes ii IsabelleVan Groeningen Thesissubmitted for the degreeof Doctorin Philosophy Universityof York Instituteof AdvancedArchitectural Studies May 1996 ST COPY AVAILA L Variable print quality Appendix 1: Summer Flowering Plants Listed by Philip Miller in 1731 APPENDIX 1: SUMMER FLOWERING PLANTS LISTED BY PHILIP MILLER IN 1731 Source: Miller, Philip: The Gardener'sDictionary, 1731 Notes: 1. The following list was published by Miller indicating what was flowering in the months of June,July, August and September,which are the four months during which the majority of herbaceousplants flower. The nomenclatureof Miller's nameshas, where possible,been updatedand addedbetween brackets with the help of Tony Lord. 2. The nomenclatureor identity of plants marked with a? is uncertain. acanthus(Acanthus spp.) aloes (Aloe vera) althaeafrutex (Hibiscus syriacus) amaranthus(Amaranthus sp. ) amaranthoides(globe amaranth:Gomphrena globosa) annual stock, July-flowers (Matthiola incana) antirrhinum or calves snouts(Antirrhinum majus), apocynum (Millees Apocynum contains severalspecies from Asclepiadeaeand Apocynaceae:Asclepias, Rhabdadenia,Echites, Forsteronia, Prestonia as well as Apocynum asters(Aster spp.) auricula,(Primula auricula) autumn hyacinth (Polyxena corymbosa) autumn crocus (Crocus speciosus) autumnalis * balsamines(Impatiens balsamina) bean caper (Zygophyllum) bloody cranesbill (Geranium sanguineum) blue featheredhyacinth (Muscari comosummonstrosum) broad-leavedupright lily of the valley (Convallaria lati/blia or Polygonatum spp) bulbous irises (Iris xiphium) bulbous fiery lily (Lilium bulbiferum) buphthalmumsof sorts (probalby Anthemis spp.) campanulas(Campanula spp.) candytuft (Iberis sempervirens) Canterbury bells (Campanula medium) Capsicum indicum (Capsicum annuum) cardinal flower (Lobelia cardinalis) carnations(Dianthus caryophyllus) catchfly (Silene diolca and S. -

Micropropagation of Tulip Via Somatic Embryogenesis
agronomy Article Micropropagation of Tulip via Somatic Embryogenesis Małgorzata Podwyszy ´nska and Agnieszka Marasek-Ciolakowska * Department of Applied Biology, Research Institute of Horticulture, Konstytucji 3 Maja 1/3 Str., 96-100 Skierniewice, Poland; [email protected] * Correspondence: [email protected] Received: 7 October 2020; Accepted: 24 November 2020; Published: 25 November 2020 Abstract: An effective method of tulip regeneration via somatic embryogenesis (SE) was developed. Explants, flower stem slices excised from cooled bulbs were incubated in darkness on MS modified media containing auxins alone (2,4-dichlorophenoxyacetic acid—2,4-D, 1-naphthalene acetic acid—NAA and 4-amino-3,5,6-trichloro-2-pyridine carboxylic acid—picloram) or combined with 1 thidiazuron (TDZ) at 0.1 and 0.5 mg L− . Yellowish-white callus with a granular structure was developed in the presence of all auxins on the cut surface from the tissues of the vascular bundles. From this, lines of embryogenic calli were derived. The addition of TDZ to the medium with auxins significantly stimulated somatic embryo formation. Cyclic and the most intensive proliferation of 1 embryogenic callus as well as embryo formation was obtained in the presence of 2,4-D at 0.1 mg L− 1 combined with TDZ at 0.5 mg L− . Addition of proline enhanced either callus proliferation rate or frequency of embryo formation. The best quality embryos with cotyledons longer than 10 mm able to form bulbs were recorded when TDZ was replaced with 6-benzylaminopurine (BAP) at the 1 concentration of 0.1 mg L− . Histomorphology showed that the development of somatic embryos could have either external or internal origins. -

Entomofauna of Pollinators Tulipa Suaveolens Roth in the Lower Volga Region
ЭНТОМОФАУНА ОПЫЛИТЕЛЕЙ TULIPA SUAVEOLENS UDC 581.5 ENTOMOFAUNA OF POLLINATORS TULIPA SUAVEOLENS ROTH IN THE LOWER VOLGA REGION Petrova N. A. 1, Kashin A. S. 1, Korneev, M. G. 2, Anikin V. V. 1 1 N. G. Chernyshevsky Saratov State University 83 Astrakhanskaya Str., Saratov 410012, Russia E-mail: [email protected] 2 Russian Research Anti-Plague Institute “Microbe” of the Rospotrebnadzor 46 Universitetskaya Str., Saratov 410005, Russia Received 1 March 2019, Accepted 25 March 2019 The list of insects collected from flowering plants of Tulipa suaveolens in twelve populations in the Saratov, Volgograd, Rostov Provinces and the Republic of Kalmykia is noted in article. Collecting was carried out by mowing with a butterfly net between еру clusters of flowering plants, as well by hand from flowers. It was established that on this largest territory the collected insects similar in faunal structure. Most of them are typical pollinators and phytophages of the steppe zone of Russia. According to results of the study, there is no reason to believe that the effect of insect pollinators on T. suaveolens can be selective in nature and have affecting on the genetic structure of populations or have the originality of the biogeographic distribution of tepals color. Key words: Tulipa suaveolens Roth, pollinators, insects, phytophages, flowering seed reproduction. DOI: 10.18500/1682-1637-2019-1-3-17 REFERENCES Anikin V. V., Saranova O. A. Spring aspect of Lepidoptera fauna (Lepidoptera) of the southern regions of the Republic of Kalmykia. In: Problems of nature management and biodiversity conservation in the conditions of desertification: materials Interregion. -

Aktualniproblemy-2017
Національна академія наук України Інститут ботаніки ім. М.Г. Холодного КТУАЛЬНІКТУАЛЬНІ ПРОБЛЕМИПРОБЛЕМИ БОТАНІКИБОТАНІКИ ТАТА ЕКОЛОГІЇЕКОЛОГІЇ матеріали міжнародної конференції молодих учених луцьк 2017 УДК 58 ББК Е52 А 43 Редакційна колегія: чл.-коР. НАН УкРаїни Єлизавета львівна коРдюм, галеб аль-маалі, денис винокуров, маРія зикова, олена білоУс, василь бРиков, надія капець, тетяна каРпюк, валеРія павленко-баРишева, олександР поліщУк, ольга чУсова Актуальні проблеми ботаніки та екології : матеріали Міжнар. А 43 конф. молодих учених (м. Луцьк, 5–10 верес. 2017 р.). – Луцьк : Вежа-Друк, 2017. – 120 с. ISBN 978-966-940-110-6 У збірнику представлено матеріали Міжнародної конференції молодих учених “Актуальні проблеми ботаніки та екології”. Висвітлено результати досліджень в галузях альгології, бріології, ліхенології, мікології, молекулярної біології, фізіології та біохімії рослин, фітогормонології, клітинної біології, генетики, анатомії, морфології та географії рослин, екології рослин, фітоценології, дендрології, інтродукції рослин, ландшафтної архітектури та ін. УДК 58 ББК Е52 ISBN 978-966-940-110-6 © Інститут ботаніки ім. М. Г. Холодного НАН України, 2017 Національна академія наук України Інститут ботаніки ім. М.Г. Холодного Східноєвропейський національний університет імені Лесі Українки Актуальні проблеми ботаніки та екології Матеріали міжнародної конференції молодих учених Луцьк – 2017 Національна академія наук України Інститут ботаніки ім. М.Г. Холодного Східноєвропейський національний університет імені Лесі Українки СЕКЦІЇ КОНФЕРЕНЦІЇ: 1. Альгологія, бріологія, ліхенологія та мікологія 2. Систематика та флористика судинних рослин 3. Екологія рослин та фітоценологія 4. Експериментальна ботаніка та мікологія 5. Дендрологія, інтродукція рослин та ландшафтна архітектура Робочі мови конференції: українська, російська, англійська Форми участі у конференції: очна (усна доповідь та постерна доповідь), заочна ОРГАНІЗАЦІЙНИЙ КОМІТЕТ КОНФЕРЕНЦІЇ Голова оргкомітету: чл.-кор. НАН України Єлизавета Львівна Кордюм (Інститут ботаніки) Співголова: к.б.н.