Quantitative Phosphoproteomic Analysis Identifies the Potential
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deregulated Gene Expression Pathways in Myelodysplastic Syndrome Hematopoietic Stem Cells
Leukemia (2010) 24, 756–764 & 2010 Macmillan Publishers Limited All rights reserved 0887-6924/10 $32.00 www.nature.com/leu ORIGINAL ARTICLE Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells A Pellagatti1, M Cazzola2, A Giagounidis3, J Perry1, L Malcovati2, MG Della Porta2,MJa¨dersten4, S Killick5, A Verma6, CJ Norbury7, E Hellstro¨m-Lindberg4, JS Wainscoat1 and J Boultwood1 1LRF Molecular Haematology Unit, NDCLS, John Radcliffe Hospital, Oxford, UK; 2Department of Hematology Oncology, University of Pavia Medical School, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy; 3Medizinische Klinik II, St Johannes Hospital, Duisburg, Germany; 4Division of Hematology, Department of Medicine, Karolinska Institutet, Stockholm, Sweden; 5Department of Haematology, Royal Bournemouth Hospital, Bournemouth, UK; 6Albert Einstein College of Medicine, Bronx, NY, USA and 7Sir William Dunn School of Pathology, University of Oxford, Oxford, UK To gain insight into the molecular pathogenesis of the the World Health Organization.6,7 Patients with refractory myelodysplastic syndromes (MDS), we performed global gene anemia (RA) with or without ringed sideroblasts, according to expression profiling and pathway analysis on the hemato- poietic stem cells (HSC) of 183 MDS patients as compared with the the French–American–British classification, were subdivided HSC of 17 healthy controls. The most significantly deregulated based on the presence or absence of multilineage dysplasia. In pathways in MDS include interferon signaling, thrombopoietin addition, patients with RA with excess blasts (RAEB) were signaling and the Wnt pathways. Among the most signifi- subdivided into two categories, RAEB1 and RAEB2, based on the cantly deregulated gene pathways in early MDS are immuno- percentage of bone marrow blasts. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Mouse Rras2 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Rras2 Knockout Project (CRISPR/Cas9) Objective: To create a Rras2 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Rras2 gene (NCBI Reference Sequence: NM_025846 ; Ensembl: ENSMUSG00000055723 ) is located on Mouse chromosome 7. 6 exons are identified, with the ATG start codon in exon 1 and the TAA stop codon in exon 6 (Transcript: ENSMUST00000069449). Exon 3~5 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Homozygote and heterozygote null mice are lymphopenic, resulting from diminished homeostatic proliferation and impaired T cell and B cell survival. Mice homozygous for a gene trap insertion exhibit retinal degeneration, and increased total body mass and total body fat. Exon 3 starts from about 32.19% of the coding region. Exon 3~5 covers 54.08% of the coding region. The size of effective KO region: ~8729 bp. The KO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 3 4 5 6 Legends Exon of mouse Rras2 Knockout region Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 1302 bp section upstream of Exon 3 is aligned with itself to determine if there are tandem repeats. Tandem repeats are found in the dot plot matrix. -

CD56+ T-Cells in Relation to Cytomegalovirus in Healthy Subjects and Kidney Transplant Patients
CD56+ T-cells in Relation to Cytomegalovirus in Healthy Subjects and Kidney Transplant Patients Institute of Infection and Global Health Department of Clinical Infection, Microbiology and Immunology Thesis submitted in accordance with the requirements of the University of Liverpool for the degree of Doctor in Philosophy by Mazen Mohammed Almehmadi December 2014 - 1 - Abstract Human T cells expressing CD56 are capable of tumour cell lysis following activation with interleukin-2 but their role in viral immunity has been less well studied. The work described in this thesis aimed to investigate CD56+ T-cells in relation to cytomegalovirus infection in healthy subjects and kidney transplant patients (KTPs). Proportions of CD56+ T cells were found to be highly significantly increased in healthy cytomegalovirus-seropositive (CMV+) compared to cytomegalovirus-seronegative (CMV-) subjects (8.38% ± 0.33 versus 3.29%± 0.33; P < 0.0001). In donor CMV-/recipient CMV- (D-/R-)- KTPs levels of CD56+ T cells were 1.9% ±0.35 versus 5.42% ±1.01 in D+/R- patients and 5.11% ±0.69 in R+ patients (P 0.0247 and < 0.0001 respectively). CD56+ T cells in both healthy CMV+ subjects and KTPs expressed markers of effector memory- RA T-cells (TEMRA) while in healthy CMV- subjects and D-/R- KTPs the phenotype was predominantly that of naïve T-cells. Other surface markers, CD8, CD4, CD58, CD57, CD94 and NKG2C were expressed by a significantly higher proportion of CD56+ T-cells in healthy CMV+ than CMV- subjects. Functional studies showed levels of pro-inflammatory cytokines IFN-γ and TNF-α, as well as granzyme B and CD107a were significantly higher in CD56+ T-cells from CMV+ than CMV- subjects following stimulation with CMV antigens. -

Euchromatin Histone Methyltransferase II (EHMT2) Regulates the Expression of Ras-Related GTP Binding C (RRAGC) Protein
BMB Rep. 2020; 53(11): 576-581 BMB www.bmbreports.org Reports Euchromatin histone methyltransferase II (EHMT2) regulates the expression of ras-related GTP binding C (RRAGC) protein Supyong Hwang1, Soyoung Kim1, Kyungkon Kim1,2,3, Jeonghun Yeom2, Sojung Park1 & Inki Kim1,2,3,* 1Biomedical Research Center, ASAN Institute for Life Sciences, ASAN Medical Center, Seoul 05505, 2Convergence Medicine Research Center (CREDIT), ASAN Institute for Life Sciences, ASAN Medical Center, Seoul 05505, 3Department of Convergence Medicine, University of Ulsan College of Medicine, Seoul 05505, Korea Dimethylation of the histone H3 protein at lysine residue 9 INTRODUCTION (H3K9) is mediated by euchromatin histone methyltransferase II (EHMT2) and results in transcriptional repression of target Epigenetic modifications are gene regulatory mechanisms that genes. Recently, chemical inhibition of EHMT2 was shown to are independent of changes in DNA sequences (1). This mode induce various physiological outcomes, including endoplasmic of gene regulation can be achieved by means of histone and reticulum stress-associated genes transcription in cancer cells. DNA modifications, such as methylation and acetylation (1). To identify genes that are transcriptionally repressed by EHMT2 Among these, methylation at lysine residues 4 (H3K4) and 36 during apoptosis, and cell stress responses, we screened genes (H3K36) of histone H3 are hallmarks of transcriptional acti- that are upregulated by BIX-01294, a chemical inhibitor of vation, whereas methylation of histone H3 residues at lysines EHMT2. RNA sequencing analyses revealed 77 genes that were 9 (H3K9) and 27 (H3K27) leads to repression (2). Methylation upregulated by BIX-01294 in all four hepatic cell carcinoma of histones is accomplished by histone methyltransferases (HMTs) (HCC) cell lines. -

Effects of Rapamycin on Social Interaction Deficits and Gene
Kotajima-Murakami et al. Molecular Brain (2019) 12:3 https://doi.org/10.1186/s13041-018-0423-2 RESEARCH Open Access Effects of rapamycin on social interaction deficits and gene expression in mice exposed to valproic acid in utero Hiroko Kotajima-Murakami1,2, Toshiyuki Kobayashi3, Hirofumi Kashii1,4, Atsushi Sato1,5, Yoko Hagino1, Miho Tanaka1,6, Yasumasa Nishito7, Yukio Takamatsu7, Shigeo Uchino1,2 and Kazutaka Ikeda1* Abstract The mammalian target of rapamycin (mTOR) signaling pathway plays a crucial role in cell metabolism, growth, and proliferation. The overactivation of mTOR has been implicated in the pathogenesis of syndromic autism spectrum disorder (ASD), such as tuberous sclerosis complex (TSC). Treatment with the mTOR inhibitor rapamycin improved social interaction deficits in mouse models of TSC. Prenatal exposure to valproic acid (VPA) increases the incidence of ASD. Rodent pups that are exposed to VPA in utero have been used as an animal model of ASD. Activation of the mTOR signaling pathway was recently observed in rodents that were exposed to VPA in utero, and rapamycin ameliorated social interaction deficits. The present study investigated the effect of rapamycin on social interaction deficits in both adolescence and adulthood, and gene expressions in mice that were exposed to VPA in utero. We subcutaneously injected 600 mg/kg VPA in pregnant mice on gestational day 12.5 and used the pups as a model of ASD. The pups were intraperitoneally injected with rapamycin or an equal volume of vehicle once daily for 2 consecutive days. The social interaction test was conducted in the offspring after the last rapamycin administration at 5–6 weeks of ages (adolescence) or 10–11 weeks of age (adulthood). -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

In Vitro Pharmacological Profiling of R406 Identifies Molecular Targets
ORIGINAL ARTICLE In vitro pharmacological profiling of R406 identifies molecular targets underlying the clinical effects of fostamatinib Michael G. Rolf, Jon O. Curwen, Margaret Veldman-Jones, Cath Eberlein, Jianyan Wang, Alex Harmer, Caroline J. Hellawell & Martin Braddock AstraZeneca R&D Alderley Park, Macclesfield, Cheshire SK10 4TG, United Kingdom Keywords Abstract Blood pressure elevation, fostamatinib, in vitro pharmacological profiling, R406, SYK Off-target pharmacology may contribute to both adverse and beneficial effects of a new drug. In vitro pharmacological profiling is often applied early in drug Correspondence discovery; there are fewer reports addressing the relevance of broad profiles to Michael G. Rolf, AstraZeneca R&D Molndal,€ clinical adverse effects. Here, we have characterized the pharmacological profile Pepparedsleden 1, 431 83 Mo¨ lndal, Sweden. of the active metabolite of fostamatinib, R406, linking an understanding of drug Tel: +46 31 776 60 40; Fax: +46 31 776 37 selectivity to the increase in blood pressure observed in clinical studies. R406 60; E-mail: [email protected] was profiled in a broad range of in vitro assays to generate a comprehensive Funding Information pharmacological profile and key targets were further investigated using func- No funding information provided. tional and cellular assay systems. A combination of traditional literature searches and text-mining approaches established potential mechanistic links Received: 9 April 2015; Accepted: 14 July between the profile of R406 and clinical side effects. R406 was selective outside 2015 the kinase domain, with only antagonist activity at the adenosine A3 receptor in the range relevant to clinical effects. R406 was less selective in the kinase Pharma Res Per, 3(5), 2015, e00175, doi: 10.1002/prp2.175 domain, having activity at many protein kinases at therapeutically relevant con- centrations when tested in multiple in vitro systems. -

Epigenetic Loss of the RNA Decapping Enzyme NUDT16 Mediates C-MYC Activation in T-Cell Acute Lymphoblastic Leukemia
OPEN Leukemia (2017) 31, 1622–1657 www.nature.com/leu LETTERS TO THE EDITOR Epigenetic loss of the RNA decapping enzyme NUDT16 mediates C-MYC activation in T-cell acute lymphoblastic leukemia Leukemia (2017) 31, 1622–1625; doi:10.1038/leu.2017.99 Having found the aforementioned NUDT16 CpG island methy- lation profiles, we studied in greater detail their association with the possible transcriptional inactivation of the NUDT16 gene at the RNA and protein levels in leukemia cell lines. We first It is possible that the occurrence of intrinsic defects in RNA performed bisulfite genomic sequencing of mutiple clones in the – processing pathways, such as RNA decapping,1 3 contribute to the T-cell Acute Lymphoblastic Leukemia (T-ALL) cell lines CCRF-CEM, distorted RNA landscapes of cancer cells. After transcription by Jurkat, MOLT-4 and MOLT-16 using primers that encompassed the RNA polymerase II, RNA molecules are equipped with a 5´-end transcription start site-associated CpG island and confirmed the N7-methyl guanosine (m7G)-cap. This m7G-cap is essential for hypermethylated status of the 5′-end region of NUDT16 in translation, stabilizing the RNA molecule and protecting it from comparison to normal T lymphocytes (Figure 1b), validating the – exonucleolytic breakdown.1 3 For RNA decay to occur the DNA methylation patterns obtained by the microarray approach m7G-cap first needs to be removed. This process is known as (Supplementary Figure S2). In contrast, normal T lymphocytes, the – decapping.1 3 The decapping mRNA 2 (DCP2) enzyme,4 also T-ALL cell lines KOPN-8, REH and RS4;11 and the leukemia cell known as the nucleoside diphosphate-linked moiety X motif lines HL-60 and K562 derived from myeloid lineage were all found 20 (NUDT20), was originally thought to be the only mammalian to be unmethylated (Figure 1b; Supplementary Figure S2). -

Phospho-RPS6KA1(T359) Antibody Peptide Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap3497a
9765 Clairemont Mesa Blvd, Suite C San Diego, CA 92124 Tel: 858.875.1900 Fax: 858.622.0609 Phospho-RPS6KA1(T359) Antibody Peptide Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP3497a Specification Phospho-RPS6KA1(T359) Antibody - Product Information Application WB, DB,E Primary Accession Q15418 Other Accession P10666, P10665, P18652 Reactivity Human Predicted Chicken, Xenopus Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 82723 Phospho-RPS6KA1(T359) Antibody - Additional Information Gene ID 6195 Other Names Western blot analysis of extracts from Hela Ribosomal protein S6 kinase alpha-1, cells,untreated or treated with S6K-alpha-1, 90 kDa ribosomal protein S6 TPA,200nM,using phospho RPS6KA1-T359 (left) kinase 1, p90-RSK 1, p90RSK1, p90S6K, MAP or RPS6KA1 antibody(right) kinase-activated protein kinase 1a, MAPK-activated protein kinase 1a, MAPKAP kinase 1a, MAPKAPK-1a, Ribosomal S6 kinase 1, RSK-1, RPS6KA1, MAPKAPK1A, RSK1 Target/Specificity This RPS6KA1 Antibody is generated from rabbits immunized with a KLH conjugated synthetic phosphopeptide corresponding to amino acid residues surrounding T359 of human RPS6KA1. Dilution WB~~1:2000 DB~~1:500 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Dot blot analysis of anti-RPS6KA1-pT359 Pab (RB13385) on nitrocellulose membrane. 50ng Storage of Phospho-peptide or Non Phospho-peptide per Maintain refrigerated at 2-8°C for up to 6 dot were adsorbed. Antibody working months. For long term storage store at -20°C concentrations are 0.5ug per ml. in small aliquots to prevent freeze-thaw cycles. -
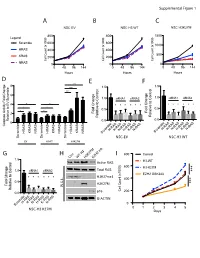
C a B D G E F
Supplemental Figure 1 A B C NSC EV NSC H3 WT NSC H3K27M 800 800 1500 ) ) Legend ) Scramble 600 600 x1000 x1000 x1000 ( ( ( 1000 t t t n n HRAS n 400 400 u u u o o o C C C 500 l l KRAS l 200 200 el el el C C C NRAS 0 0 0 0 48 96 144 0 48 96 144 0 48 96 144 Hours Hours Hours D *** E F 8 1.5 1.5 e *** l l *** ro ro t 6 t 1.0 siRNA1 siRNA2 1.0 siRNA1 siRNA2 Con Con o o t 4 * * t * * * * * * e * * * * * * e v i * * v i t 0.5 * * t 0.5 Fold Change 2 Fold Change Rela Rela Relative to EV Scrambl 0.0 0.0 0 Caspase Activity Fold Change H-RASK-RASN-RASH-RASK-RASN-RAS H-RASK-RASN-RASH-RASK-RASN-RAS KRAS KRAS KRAS HRAS NRAS HRAS NRAS HRAS NRAS Scramble Scramble Scramble Scramble Scramble NSC-EV NSC-H3 WT EV H3WT H3K27M G H I 800 Control Con WT H3 H3K27MEZH2 inb 1.5 l H3-WT Active RAS ro ) t 600 H3-K27M 1.0 siRNA1 siRNA2 Total RAS *** Con x1000 ( o EZH2 GSK343 t t * * * * * * H3K27me3 n e 400 u v i o t 0.5 *** H3K27M C Fold Change l W.C.L. el Rela p16 C 200 0.0 B-ACTIN H-RASK-RASN-RASH-RASK-RASN-RAS 0 Scramble NSC-H3 K27M 0 1 2 3 4 5 Days E C A 100 Cell Viability 100 25 50 75 Cell Viability 25 50 75 0 0 Fold Change MOCK MOCK Relative to Control Scramble * 0.0 0.2 0.4 0.6 0.8 1.0 1.2 MYC Scramble MYC MYC * PDGFRA Scramble siRNA PDGFRA MYC PDGFRA AURKA AURKA DIPG007siRNA2 PDGFRA NSC H3K27MsiRNA1and AURKA LAMTOR3 AURKA LAMTOR3 LAMTOR3 LIN28A LAMTOR3 LIN28A LIN28B LIN28A NSC-EV LIN28A MAP2K3 LIN28B LIN28B LIN28B MAP2K5 siRNA1 MAP2K3 MAP2K3 MAP3K2 MAP2K3 MAP3K7 MAP2K5 MAP2K5 MAPK7 MAP2K5 2 MAP3K2 siRNA1 siRNA2 MAP3K2 ZAK MAP3K7 MAP3K2 MAP3K7 MAPK7 MAP3K7 -

Novel Polymorphisms in RAPGEF6 Gene Associated with Egg-Laying Rate in Chinese Jing Hong Chicken Using Genome-Wide SNP Scan
G C A T T A C G G C A T genes Article Novel Polymorphisms in RAPGEF6 Gene Associated with Egg-Laying Rate in Chinese Jing Hong Chicken using Genome-Wide SNP Scan Syed Ali Azmal 1,2, Ali Akbar Bhuiyan 1,3, Abdullah Ibne Omar 1, Shuai Ma 1, Chenghao Sun 4, Zhongdong Han 4, Meikuen Zhang 5, Shuhong Zhao 1 and Shijun Li 1,* 1 Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction, Ministry of Education, College of Animal Science and Veterinary Medicine, Huazhong Agricultural University, Wuhan 430070, China; [email protected] (S.A.A.); [email protected] (A.A.B.); [email protected] (A.I.O.); [email protected] (S.M.); [email protected] (S.Z.) 2 Department of Livestock Services (DLS), Under the Ministry of Fisheries and Livestock (MOFL), Dhaka 1000, Bangladesh 3 Biotechnology Division, Bangladesh Livestock Research Institute, Under the Ministry of Fisheries and Livestock (MOFL), Dhaka 1000, Bangladesh 4 Huadu Yukou Poultry Industry Co. Ltd., Beijing 100000, China; [email protected] (C.S.); [email protected] (Z.H.) 5 DQY Ecological Co. Ltd., Beijing 100000, China; [email protected] * Correspondence: [email protected]; Tel.: +86-27-87281306; Fax: +86-27-87280408 Received: 4 April 2019; Accepted: 14 May 2019; Published: 20 May 2019 Abstract: The improvement of egg production is of vital importance in the chicken industry to maintain optimum output throughout the laying period. Because of the elongation of the egg-laying cycle, a drop in egg-laying rates in the late laying period has provoked great concern in the poultry industry.