Evolution of Proteins Involved in Response To
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
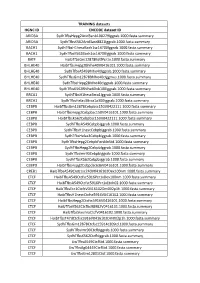
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

The Autophagy Receptor SQSTM1/P62 Mediates Anti-Inflammatory Actions of the Selective NR3C1/ Glucocorticoid Receptor Modulator Compound a (Cpda) in Macrophages
Autophagy ISSN: 1554-8627 (Print) 1554-8635 (Online) Journal homepage: http://www.tandfonline.com/loi/kaup20 The autophagy receptor SQSTM1/p62 mediates anti-inflammatory actions of the selective NR3C1/ glucocorticoid receptor modulator compound A (CpdA) in macrophages Viacheslav Mylka, Julie Deckers, Dariusz Ratman, Lode De Cauwer, Jonathan Thommis, Riet De Rycke, Francis Impens, Claude Libert, Jan Tavernier, Wim Vanden Berghe, Kris Gevaert & Karolien De Bosscher To cite this article: Viacheslav Mylka, Julie Deckers, Dariusz Ratman, Lode De Cauwer, Jonathan Thommis, Riet De Rycke, Francis Impens, Claude Libert, Jan Tavernier, Wim Vanden Berghe, Kris Gevaert & Karolien De Bosscher (2018) The autophagy receptor SQSTM1/p62 mediates anti- inflammatory actions of the selective NR3C1/glucocorticoid receptor modulator compound A (CpdA) in macrophages, Autophagy, 14:12, 2049-2064, DOI: 10.1080/15548627.2018.1495681 To link to this article: https://doi.org/10.1080/15548627.2018.1495681 © 2018 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group. Published online: 14 Sep 2018. Submit your article to this journal Article views: 907 View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=kaup20 AUTOPHAGY 2018, VOL. 14, NO. 12, 2049–2064 https://doi.org/10.1080/15548627.2018.1495681 RESEARCH PAPER - BASIC SCIENCE The autophagy receptor SQSTM1/p62 mediates anti-inflammatory actions of the selective NR3C1/glucocorticoid receptor modulator -

Expression Profiling of KLF4
Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in wild type MEFs. 98 Am J Cancer Res 2011;1(1):85-97 Table S1: Functional Annotation Clustering of Genes Up-Regulated in Klf4 -Null MEFs ILLUMINA_ID Gene Symbol Gene Name (Description) P -value Fold-Change Cell Cycle 8.00E-03 ILMN_1217331 Mcm6 MINICHROMOSOME MAINTENANCE DEFICIENT 6 40.36 ILMN_2723931 E2f6 E2F TRANSCRIPTION FACTOR 6 26.8 ILMN_2724570 Mapk12 MITOGEN-ACTIVATED PROTEIN KINASE 12 22.19 ILMN_1218470 Cdk2 CYCLIN-DEPENDENT KINASE 2 9.32 ILMN_1234909 Tipin TIMELESS INTERACTING PROTEIN 5.3 ILMN_1212692 Mapk13 SAPK/ERK/KINASE 4 4.96 ILMN_2666690 Cul7 CULLIN 7 2.23 ILMN_2681776 Mapk6 MITOGEN ACTIVATED PROTEIN KINASE 4 2.11 ILMN_2652909 Ddit3 DNA-DAMAGE INDUCIBLE TRANSCRIPT 3 2.07 ILMN_2742152 Gadd45a GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE 45 ALPHA 1.92 ILMN_1212787 Pttg1 PITUITARY TUMOR-TRANSFORMING 1 1.8 ILMN_1216721 Cdk5 CYCLIN-DEPENDENT KINASE 5 1.78 ILMN_1227009 Gas2l1 GROWTH ARREST-SPECIFIC 2 LIKE 1 1.74 ILMN_2663009 Rassf5 RAS ASSOCIATION (RALGDS/AF-6) DOMAIN FAMILY 5 1.64 ILMN_1220454 Anapc13 ANAPHASE PROMOTING COMPLEX SUBUNIT 13 1.61 ILMN_1216213 Incenp INNER CENTROMERE PROTEIN 1.56 ILMN_1256301 Rcc2 REGULATOR OF CHROMOSOME CONDENSATION 2 1.53 Extracellular Matrix 5.80E-06 ILMN_2735184 Col18a1 PROCOLLAGEN, TYPE XVIII, ALPHA 1 51.5 ILMN_1223997 Crtap CARTILAGE ASSOCIATED PROTEIN 32.74 ILMN_2753809 Mmp3 MATRIX METALLOPEPTIDASE -

Roles of NRF3 in the Hallmarks of Cancer: Proteasomal Inactivation of Tumor Suppressors
cancers Review Roles of NRF3 in the Hallmarks of Cancer: Proteasomal Inactivation of Tumor Suppressors Akira Kobayashi 1,2 1 Laboratory for Genetic Code, Graduate School of Life and Medical Sciences, Doshisha University, Kyotanabe, Kyoto 610-0394, Japan; [email protected]; Tel.: +81-774-65-6273 2 Department of Medical Life Systems, Faculty of Life and Medical Sciences, Doshisha University, Kyotanabe, Kyoto 610-0394, Japan Received: 1 September 2020; Accepted: 17 September 2020; Published: 20 September 2020 Simple Summary: This review summarizes recent advances in our understanding of the physiological roles of the NFE2-related factor 2 (NRF2)-related transcription factor NRF3 in cancer. NRF3 confers cells with six so-called “hallmarks of cancer” through upregulating gene expression of specific target genes, leading to tumorigenesis and cancer malignancy. These driver gene-like functions of NRF3 in cancer are distinct from those of NRF2. Abstract: The physiological roles of the NRF2-related transcription factor NRF3 (NFE2L3) have remained unknown for decades. The remarkable development of human cancer genome databases has led to strong suggestions that NRF3 has functional significance in cancer; specifically,high NRF3 mRNA levels are induced in many cancer types, such as colorectal cancer and pancreatic adenocarcinoma, and are associated with poor prognosis. On the basis of this information, the involvement of NRF3 in tumorigenesis and cancer malignancy has been recently proposed. NRF3 confers cancer cells with selective growth advantages by enhancing 20S proteasome assembly through induction of the chaperone gene proteasome maturation protein (POMP) and consequently promoting degradation of the tumor suppressors p53 and retinoblastoma (Rb) in a ubiquitin-independent manner. -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

Genome-Scale Identification of Transcription Factors That Mediate An
ARTICLE DOI: 10.1038/s41467-018-04406-2 OPEN Genome-scale identification of transcription factors that mediate an inflammatory network during breast cellular transformation Zhe Ji 1,2,4, Lizhi He1, Asaf Rotem1,2,5, Andreas Janzer1,6, Christine S. Cheng2,7, Aviv Regev2,3 & Kevin Struhl 1 Transient activation of Src oncoprotein in non-transformed, breast epithelial cells can initiate an epigenetic switch to the stably transformed state via a positive feedback loop that involves 1234567890():,; the inflammatory transcription factors STAT3 and NF-κB. Here, we develop an experimental and computational pipeline that includes 1) a Bayesian network model (AccessTF) that accurately predicts protein-bound DNA sequence motifs based on chromatin accessibility, and 2) a scoring system (TFScore) that rank-orders transcription factors as candidates for being important for a biological process. Genetic experiments validate TFScore and suggest that more than 40 transcription factors contribute to the oncogenic state in this model. Interestingly, individual depletion of several of these factors results in similar transcriptional profiles, indicating that a complex and interconnected transcriptional network promotes a stable oncogenic state. The combined experimental and computational pipeline represents a general approach to comprehensively identify transcriptional regulators important for a biological process. 1 Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA. 2 Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA. 3 Department of Biology, Howard Hughes Medical Institute and David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 20140, USA. 4Present address: Department of Pharmacology and Biomedical Engineering, Northwestern University, Evanston 60611 IL, USA. -

NFE2L1 Antibody Cat
NFE2L1 Antibody Cat. No.: 31-335 NFE2L1 Antibody Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human, Mouse, Rat Antibody produced in rabbits immunized with a synthetic peptide corresponding a region IMMUNOGEN: of human NFE2L1. TESTED APPLICATIONS: ELISA, WB NFE2L1 antibody can be used for detection of NFE2L1 by ELISA at 1:7862500. NFE2L1 APPLICATIONS: antibody can be used for detection of NFE2L1 by western blot at 1 μg/mL, and HRP conjugated secondary antibody should be diluted 1:50,000 - 100,000. POSITIVE CONTROL: 1) Cat. No. 1205 - Jurkat Cell Lysate PREDICTED MOLECULAR 85 kDa WEIGHT: Properties PURIFICATION: Antibody is purified by peptide affinity chromatography method. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid September 27, 2021 1 https://www.prosci-inc.com/nfe2l1-antibody-31-335.html Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% BUFFER: sucrose. CONCENTRATION: batch dependent For short periods of storage (days) store at 4˚C. For longer periods of storage, store STORAGE CONDITIONS: NFE2L1 antibody at -20˚C. As with any antibody avoid repeat freeze-thaw cycles. Additional Info OFFICIAL SYMBOL: NFE2L1 ALTERNATE NAMES: NFE2L1, FLJ00380, LCR-F1, NRF1, TCF11 ACCESSION NO.: NP_003195 PROTEIN GI NO.: 4505379 GENE ID: 4779 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References NFE2L1 activates erythroid-specific, globin gene expression. This gene encodes a protein that is involved in globin gene expression in erythrocytes. Confusion has occurred in bibliographic databases due to the shared symbol of NRF1 for this gene, NFE2L1, and for BACKGROUND: 'nuclear respiratory factor 1' which has an official symbol of NRF1. -

(12) Patent Application Publication (10) Pub. No.: US 2006/0068395 A1 Wood Et Al
US 2006.0068395A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2006/0068395 A1 Wood et al. (43) Pub. Date: Mar. 30, 2006 (54) SYNTHETIC NUCLEIC ACID MOLECULE (21) Appl. No.: 10/943,508 COMPOSITIONS AND METHODS OF PREPARATION (22) Filed: Sep. 17, 2004 (76) Inventors: Keith V. Wood, Mt. Horeb, WI (US); Publication Classification Monika G. Wood, Mt. Horeb, WI (US); Brian Almond, Fitchburg, WI (51) Int. Cl. (US); Aileen Paguio, Madison, WI CI2O I/68 (2006.01) (US); Frank Fan, Madison, WI (US) C7H 2L/04 (2006.01) (52) U.S. Cl. ........................... 435/6: 435/320.1; 536/23.1 Correspondence Address: SCHWEGMAN, LUNDBERG, WOESSNER & (57) ABSTRACT KLUTH 1600 TCF TOWER A method to prepare synthetic nucleic acid molecules having 121 SOUTHEIGHT STREET reduced inappropriate or unintended transcriptional charac MINNEAPOLIS, MN 55402 (US) teristics when expressed in a particular host cell. Patent Application Publication Mar. 30, 2006 Sheet 1 of 2 US 2006/0068395 A1 Figure 1 Amino Acid Codon Phe UUU, UUC Ser UCU, UCC, UCA, UCG, AGU, AGC Tyr UAU, UAC Cys UGU, UGC Leu UUA, UUG, CUU, CUC, CUA, CUG Trp UGG Pro CCU, CCC, CCA, CCG His CAU, CAC Arg CGU, CGC, CGA, CGG, AGA, AGG Gln CAA, CAG Ile AUU, AUC, AUA Thr ACU, ACC, ACA, ACG ASn AAU, AAC LyS AAA, AAG Met AUG Val GUU, GUC, GUA, GUG Ala GCU, GCC, GCA, GCG Asp GAU, GAC Gly GGU, GGC, GGA, GGG Glu GAA, GAG Patent Application Publication Mar. 30, 2006 Sheet 2 of 2 US 2006/0068395 A1 Spd Sequence pGL4B-4NN3. -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

JASPAR 2014: an Extensively Expanded and Updated Open-Access Database of Transcription Factor Binding Profiles
JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Anthony Mathelier, Xiaobei Zhao, Allen W Zhang, François Parcy, Rebecca Worsley-Hunt, David J Arenillas, Sorana Buchman, Chih-Yu Chen, Alice Chou, Hans Ienasescu, et al. To cite this version: Anthony Mathelier, Xiaobei Zhao, Allen W Zhang, François Parcy, Rebecca Worsley-Hunt, et al.. JASPAR 2014: an extensively expanded and updated open-access database of transcription fac- tor binding profiles.. Nucleic Acids Research, Oxford University Press, 2014, 42 (1), pp.D142-7. 10.1093/nar/gkt997. hal-00943558 HAL Id: hal-00943558 https://hal.archives-ouvertes.fr/hal-00943558 Submitted on 28 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial| 4.0 International License D142–D147 Nucleic Acids Research, 2014, Vol. 42, Database issue Published online 4 November 2013 doi:10.1093/nar/gkt997 JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles Anthony Mathelier1, Xiaobei Zhao2,3, Allen W. Zhang1, Franc¸ ois Parcy4, Rebecca Worsley-Hunt1, David J. Arenillas1, Sorana Buchman2, Chih-yu Chen1, Alice Chou1, Hans Ienasescu2, Jonathan Lim1, Casper Shyr1, Ge Tan4, Michelle Zhou1, Boris Lenhard5,6,*, Albin Sandelin2,* and Wyeth W. -

Supplementary Table 5. Clover Results Indicate the Number Of
Supplementary Table 5. Clover results indicate the number of chromosomes with transcription factor binding motifs statistically over‐ or under‐represented in HTE DHS within intergenic sequence (more than 2kb outside of any gene). Analysis was divided into three groups (all DHS, HTE‐selective DHS, and ubiquitous DHS). Motifs with more than one entry in the databases utilized were edited to retain only the first occurrence of the motif. All DHS x Intergenic TEselective DHS x Intergenic Ubiquitous DHS x Intergenic ID Name p < 0.01 p > 0.99 ID Name p < 0.01 p > 0.99 ID Name p < 0.01 p > 0.99 MA0002.2 RUNX1 23 0 MA0080.2 SPI1 23 0 MA0055.1 Myf 23 0 MA0003.1 TFAP2A 23 0 MA0089.1 NFE2L1::MafG 23 0 MA0068.1 Pax4 23 0 MA0039.2 Klf4 23 0 MA0098.1 ETS1 23 0 MA0080.2 SPI1 23 0 MA0055.1 Myf 23 0 MA0099.2 AP1 23 0 MA0098.1 ETS1 23 0 MA0056.1 MZF1_1‐4 23 0 MA0136.1 ELF5 23 0 MA0139.1 CTCF 23 0 MA0079.2 SP1 23 0 MA0145.1 Tcfcp2l1 23 0 V$ALX3_01 ALX‐3 23 0 MA0080.2 SPI1 23 0 MA0150.1 NFE2L2 23 0 V$ALX4_02 Alx‐4 23 0 myocyte enhancer MA0081.1 SPIB 23 0 MA0156.1 FEV 23 0 V$AMEF2_Q6 factor 23 0 MA0089.1 NFE2L1::MafG 23 0 V$AP1FJ_Q2 activator protein 1 23 0 V$AP1_01 AP‐1 23 0 MA0090.1 TEAD1 23 0 V$AP4_Q5 activator protein 4 23 0 V$AP2_Q6_01 AP‐2 23 0 MA0098.1 ETS1 23 0 V$AR_Q6 half‐site matrix 23 0 V$ARX_01 Arx 23 0 BTB and CNC homolog MA0099.2 AP1 23 0 V$BACH1_01 1 23 0 V$BARHL1_01 Barhl‐1 23 0 BTB and CNC homolog MA0136.1 ELF5 23 0 V$BACH2_01 2 23 0 V$BARHL2_01 Barhl2 23 0 MA0139.1 CTCF 23 0 V$CMAF_02 C‐MAF 23 0 V$BARX1_01 Barx1 23 0 MA0144.1 Stat3 23 0