Disanthus Cercidifolius Subsp. Longipes (Hamamelidaceae) Based on AFLP Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NEWSLETTER 140, January 2018
No. 140 Irish Garden Plant Society Newsletter January 2018 Irish Heritage Daffodils Irish Heritage Daffodils IGPS Newsletter January 2018 Irish Heritage Daffodils Editorial Irish Heritage Daffodils Irish Heritage Daffodils Narcissus ‘Border Beauty’ Mary Montaut, Leinster Branch IGPS Narcissus ‘Border Beauty’ The chilly, bright winter weather just after Christmas made me really appreciate some scented subjects in the garden, especially my favourite Daphne bholua ‘Jacqueline Postil’. I was extremely fortunate many years ago to attend a propagation workshop with IGPS at Kinsealy, and they had rooted cuttings of this glorious plant. I bought one and I have adored it ever since. However, I have recently fallen in love just as passionately with another winter-scented shrub and this one, I believe, might be adopted by IGPS as one of our ‘Irish Heritage’ plants, because it is named after an Irish botanist. The shrub is Edgeworthia chrysantha - the golden-headed Edgeworthia. It belongs to the same family as the Daphne, Thymelaeaceae, and originates from the China - Nepal border area. It is naturalized in Japan, where it was planted in the late sixteenth century for paper making and is called the Paper Bush (Mitsumata). There is also an orange- flowered variety called Akebono which is said to be a smaller shrub, but I have never be lucky enough to see this one. It was first classified in 1841, and named in honour Michael Pakenham Edgeworth. He was a younger brother of the novelist, Maria Edgeworth (of Castle Rackrent fame) and lived and worked in India most of his life. However, I feel we should salute his work, and recommend this superb and tolerant shrub. -
Botanical Name Common Name
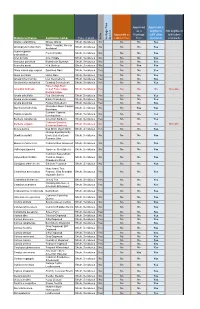
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Reproductive Biology of the Rare Plant, Dysosma Pleiantha (Berberidaceae): Breeding System, Pollination and Implications for Conservation
Pak. J. Bot ., 47(3): 951-957, 2015. REPRODUCTIVE BIOLOGY OF THE RARE PLANT, DYSOSMA PLEIANTHA (BERBERIDACEAE): BREEDING SYSTEM, POLLINATION AND IMPLICATIONS FOR CONSERVATION XI GONG 1, BI-CAI GUAN 2, *, SHI-LIANG ZHOU 3 AND GANG GE 2 1State Key Laboratory of Food Science and Technology, College of Life Science and Food engineering, Nanchang University, Nanchang 330047, China 2Jiangxi Key Laboratory of Plant Resources, Nanchang University, Nanchang 330031, China. 3State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China. *Corresponding author e-mail: [email protected], Tel.: +86 0791 83969530) Abstract Dysosma pleiantha is an endangered and endemic species in China. We have reported the flowering phenology, breeding system and pollinator activity of the species distributed in Tianmu Mountain (Zhejiang Province) nature reserves. Flowering occurred during the months of early April to late May, with the peak in the middle of the April, and was synchronous across all four subpopulations. The anthesis of an intact inflorescence lasted from sixteen to twenty-three days with eight to eleven days blossom of an individual flower. In D. pleiantha , the morphological development of flowers and fruit leading to the development of mature seeds takes place over a period 3–5 months from flowering. The average of pollen-ovule ratio (P/O) was 18 898.7. The pollen transfer in this species was mainly performed by flies, Hydrotaea chalcogaster (Muscidae). Controlled pollination experiments indicated D. pleiantha was obligate xenogamyous and self- incompatible, and pollination was pollinator-dependent. Controlled pollination experiments showed that the mean fruit set (%) under the natural condition (17.1%) was markedly lower than that of manual cross-pollination (75.6%). -

Annual Benefit Plant Sale 2012
Annual Benefit Plant Sale 2012 Botanic Gardens COLLEGE OF AGRICULTURE & NATURAL RESOURCES Connect to nature Get inspired by wildflowers, naturalistic gardening and meadows in a whole new way with our seasonal garden tours. Enjoy an art class in the garden or learn about native plant gardening, conservation, and habitats by taking one of our classes. And if you can’t visit us, enroll in our new online distance learning program, Mt. Cuba Center Connect. Visit www.mtcubacenter.org to reserve a tour or sign up for a class. Two-Hour Guided Tours | $5 per person Spring Wildflower Tours April 12th – May 27th Summer Twilight Tours May 30th – July 26th 8th Annual Wildflower Celebration |Free th April 29 , 10am – 4pm Purple pitcherplant (Sarracenia purpurea) Greenville, DE P: 302.239.4244 www.mtcubacenter.org INSPIRATION x EDUCATION x CONSERVATION 2 2012 SPRING PLANT SALE CATALOG WEBSITE: http://ag.udel.edu/udbg/events/annualsale.html WELCOME We welcome you to the twentieth annual UDBG benefit plant sale. In addition to its role as the major source of funding for the UDBG, 2012 BENEFIT PLANT SALE CATALOG we hope it also serves as a major educational event for our members and the public. It presents an opportunity to learn about new plants and consider possibilities. We should always look for ways to expand and improve our knowledge about plants and this catalog offers possibilities to accomplish both. As always, we offer an in- depth look at a particular group of plants, this year the genus Camellia. The selection goes beyond offering various cultivars with differing flower color, to a more extensive exploration of the genus with particular emphasis on hardy selections and new hybrids Camellia ‘Autumn Spirit’. -

P. Aswathi.Cdr
T REPRO N DU LA C The International Journal of Plant Reproductive Biology 10(1) Jan., 2018, pp.65-68 P T I F V O E Y B T I DOI 10.14787/ijprb.2018 10.1.65-68 O E I L O C G O S I S E T S H Reproductive biology of Malabar tamarind (Garcinia gummi-gutta (L.) T Rob.: An endemic, medicinal and spice plant from Western Ghats P. Aswathi, K. Aswani and M. Sabu* Taxonomy Division, Department of Botany, University of Calicut, Kerala - 673 635, India *e-mail : [email protected] Received : 20.07.2017; Revised: 08.10.2017; Accepted and Published online: 01.01.2018 ABSTRACT Garcinia gummi-gutta, commonly called as Malabar tamarind or Kudampuli, is endemic to Western Ghats in India. The plant is a good source of medicinally important phytochemicals. Restricted distribution, habitat destruction and over harvesting for medicinal uses leads to declining of G. gummi-gutta population. Reproductive biology studies such as flowering phenology, flower morphology, pollen viability, stigma receptivity and pollination mechanism were carried out to suggest conservation measures. Garcinia gummi-gutta is dioecious, and the flowering season for both, male and female plants, occurred in March- April months. Anthesis of both male and female flowers and stigma receptivity were observed at evening hours. Small sized pollen grains were spherical in shape and bi, tetra or penta porate. About 4230 ± 602.7 pollen grains were produced per flower. On the day of anthesis high percentage of pollen grains were fertile. Stigma is wet type and broad umbrella-shaped. -

Botany-Illustrated-J.-Glimn-Lacy-P.-Kaufman-Springer-2006.Pdf
Janice Glimn-Lacy Peter B. Kaufman 6810 Shadow Brook Court Department of Molecular, Cellular, and Indianapolis, IN 46214-1901 Developmental Biology USA University of Michigan [email protected] Ann Arbor, MI 48109-1048 USA [email protected] Library of Congress Control Number: 2005935289 ISBN-10: 0-387-28870-8 eISBN: 0-387-28875-9 ISBN-13: 978-0387-28870-3 Printed on acid-free paper. C 2006 Janice Glimn-Lacy and Peter B. Kaufman All rights reserved. This work may not be translated or copied in whole or in part without the written permission of the publisher (Springer Science+Business Media, Inc., 233 Spring Street, New York, NY 10013, USA), except for brief excerpts in connection with reviews or scholarly analysis. Use in connection with any form of information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed is forbidden. The use in this publication of trade names, trademarks, service marks, and similar terms, even if they are not identified as such, is not to be taken as an expression of opinion as to whether or not they are subject to proprietary rights. Printed in the United States of America. (TB/MVY) 987654321 springer.com Preface This is a discovery book about plants. It is for everyone For those interested in the methods used and the interested in plants including high school and college/ sources of plant materials in the illustrations, an expla- university students, artists and scientific illustrators, nation follows. For a developmental series of drawings, senior citizens, wildlife biologists, ecologists, profes- there are several methods. -

Dysosma Versipellis in PC3 and Bcap-37 Cell Lines Xiaoqiang Xu, Xiuhong Gao, Linhong Jin, Pinaki S Bhadury, Kai Yuan, Deyu Hu, Baoan Song* and Song Yang*
Xu et al. Cell Division 2011, 6:14 http://www.celldiv.com/content/6/1/14 RESEARCH Open Access Antiproliferation and cell apoptosis inducing bioactivities of constituents from Dysosma versipellis in PC3 and Bcap-37 cell lines Xiaoqiang Xu, Xiuhong Gao, Linhong Jin, Pinaki S Bhadury, Kai Yuan, Deyu Hu, Baoan Song* and Song Yang* Abstract Background: Recently, interest in phytochemicals from traditional Chinese medicinal herbs with the capability to inhibit cancer cells growth and proliferation has been growing rapidly due to their nontoxic nature. Dysosma versipellis as Bereridaceae plants is an endemic species in China, which has been proved to be an important Chinese herbal medicine because of its biological activity. However, systematic and comprehensive studies on the phytochemicals from Dysosma versipellis and their bioactivity are limited. Results: Fifteen compounds were isolated and characterized from the roots of Dysosma versipellis, among which six compounds were isolated from this plant for the first time. The inhibitory activities of these compounds were investigated on tumor cells PC3, Bcap-37 and BGC-823 in vitro by MTT method, and the results showed that podophyllotoxone (PTO) and 4’-demethyldeoxypodophyllotoxin (DDPT) had potent inhibitory activities against the growth of human carcinoma cell lines. Subsequent fluorescence staining and flow cytometry analysis indicated that these two compounds could induce apoptosis in PC3 and Bcap-37 cells, and the apoptosis ratios reached the peak (12.0% and 14.1%) after 72 h of treatment at 20 μM, respectively. Conclusions: This study suggests that most of the compounds from the roots of D. versipellis could inhibit the growth of human carcinoma cells. -

Changes in Leaf Functional Traits of the Endangered Plant Disanthus Cercidifolius Var
Changes in leaf functional traits of the endangered plant Disanthus cercidifolius var. longipes with plant growth and development Fang-Qing Chen Chemistry and Life Science College https://orcid.org/0000-0002-0724-6404 Shoupeng Guan China Three Gorges University Mengya Geng China Three Gorges University Kun Lv China Three Gorges University Yongwen Huang ( [email protected] ) Research article Keywords: Disanthus cercidifolius var. longipes, leaf functional traits, developmental stages, economic strategy Posted Date: March 18th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-17899/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/12 Abstract Background: Disanthus cercidifolius var. longipes H. T. Chang is a rare and endangered plant distributed only in the high mountains of southeastern China. In order to reveal the variation in leaf functional traits and plant investment strategies with the change of growth and developmental stages of this species, the leaf functional traits and the trait syndrome including leaf thickness (LT), leaf area (LA), leaf water content (LWC), specic leaf area (SLA), leaf total nitrogen content (LNC), and leaf total phosphorus content (LPC) of plants at different growth and developmental stages were investigated. Results:The leaf functional traits of the plants signicantly differed at different developmental stages. LT and LA of the plants increased during growth and development. LT and LA of the adult plants were 36.65% and 84.23% higher than those of the seedlings, respectively. In contrast, SLA, LWC, LNC, and LPC decreased, and in adult plants they were 48.91%, 6.63%, 8.49%, and 34.66% lower, respectively, than in seedlings. -

Tree of the Year : Parrotia the Genus Parrotia Has Two Species: P
Tree of the Year : Parrotia The genus Parrotia has two species: P. persica, familiar to dendrologists, is very variable both in the wild and in cultivation; P. subaequalis is much rarer. SUSYN ANDREWS* has been researching these fascinating plants – with contributions from Colin Crosbie, Peter Del Tredici, Michael Dosmann, Gert Fortgens, Roy Lancaster and especially Mikinori Ogisu. One of the joys of putting together the “Trees of the Year” since 1993 is that you never know what gems of information you are going to unearth, whether on a letter attached to a herbarium specimen, a little known paper in an obscure journal, an observation on an herbarium label or a chance remark by someone in passing, will lead you down a convoluted trail of discovery. The highlight for this year has been the amassing of everything I could find on the enigmatic Parrotia subaequalis, the discovery of which has had dendrologists champing at the bit to possess a plant, myself included! 6 Introduction The Hamamelidaceae is one of the key areas for palaeobotanists and taxonomists to study the evolutionary history of the Hamamelids, especially the higher taxa. In his account of the family, H. Harms (1930) provided a comprehensive suprageneric classification for the Hamamelidaceae, recognising five subfamilies, Disanthoideae (1 genus), Hamamelidoideae (16 genera), Rhodoleioideae (1 genus), Bucklandioideae (1 genus) and Liquidambaroideae (2 genera). Although a shorter treatment was published by Schulze-Menze (1964), most workers still followed Harms. Since the mid-1960s, however, much new knowledge on the family has been amassed and this has led to new thinking on the intrafamilial relationships. -

Natural Selection and Repeated Patterns of Molecular Evolution
RESEARCH ARTICLE Natural selection and repeated patterns of molecular evolution following allopatric divergence Yibo Dong1,2†, Shichao Chen3,4,5†, Shifeng Cheng6, Wenbin Zhou1, Qing Ma1, Zhiduan Chen7, Cheng-Xin Fu8, Xin Liu6*, Yun-peng Zhao8*, Pamela S Soltis3*, Gane Ka-Shu Wong6,9,10*, Douglas E Soltis3,4*, Qiu-Yun(Jenny) Xiang1* 1Department of Plant and Microbial Biology, North Carolina State University, Raleigh, United States; 2Plant Biology Division, Noble Research Institute, Ardmore, United States; 3Florida Museum of Natural History, University of Florida, Gainesville, United States; 4Department of Biology, University of Florida, Gainesville, United States; 5School of Life Sciences and Technology, Tongji University, Shanghai, China; 6Beijing Genomics Institute, Shenzhen, China; 7State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China; 8Laboratory of Systematic & Evolutionary Botany and Biodiversity, College of Life Sciences, Zhejiang University, Hangzhou, China; 9Department of Biological Sciences, University of Alberta, Edmonton, Canada; 10Department of Medicine, University of Alberta, Edmonton, Canada *For correspondence: [email protected] (XL); Abstract Although geographic isolation is a leading driver of speciation, the tempo and pattern [email protected] (Y-Z); of divergence at the genomic level remain unclear. We examine genome-wide divergence of [email protected] (PSS); putatively single-copy orthologous genes (POGs) in 20 allopatric species/variety pairs from diverse [email protected] (GK-SW); angiosperm clades, with 16 pairs reflecting the classic eastern Asia-eastern North America floristic [email protected] (DES); disjunction. In each pair, >90% of POGs are under purifying selection, and <10% are under positive [email protected] (Q-Y(J)X) selection. -

Towards a Floristic Inventory of Bat Xat Nature Reserve, Vietnam
Wulfenia 27 (2020): 233 –250 Mitteilungen des Kärntner Botanikzentrums Klagenfurt Towards a floristic inventory of Bat Xat Nature Reserve, Vietnam: Thirteen new national records of vascular plants Bui Hong Quang, Tran The Bach, Sangmi Eum, Do Van Hai, Nguyen Sinh Khang, Le Ngoc Han, Tran Duc Binh, Nguyen Thu Thuy, Vu Anh Thuong, Ngo Kien Trung, Ya-Ping Chen, Peter W. Fritsch, Chi-Ming Hu, Lu Thi Ngan, John A. N. Parnell, Alexander N. Sennikov, John R. I. Wood, Yi Yang, Andrey N. Kuznetsov, Svetlana P. Kuznetsova & Maxim S. Nuraliev Summary: The present study reports newly recorded species of vascular plants for the flora of Vietnam found in the recently established Bat Xat Nature Reserve in Lao Cai province, close to the border with Yunnan province of China. Thirteen species belonging to eleven families are reported: Acanthaceae (Strobilanthes helicta), Actinidiaceae (Actinidia melliana), Amaryllidaceae (Allium wallichii), Aquifoliaceae (Ilex fragilis), Asteraceae (Melanoseris leiolepis), Begoniaceae (Begonia yuii ), Lamiaceae (Callicarpa giraldii, Clerodendrum peii, Scutellaria macrosiphon), Lentibulariaceae (Utricularia spinomarginata), Primulaceae (Lysimachia septemfida), Pteridaceae (Aleuritopteris chrysophylla) and Symplocaceae (Symplocos glandulifera). Some of these species are additionally reported from the neighbouring Hoang Lien National Park. For each species, information on its habitat, phenology, distribution and studied specimens is provided along with the photographs of the reported findings. Keywords: Hoang Lien National Park, Indochinese Peninsula, Lao Cai province, Southeast Asia, Vietnam-China border area, Y Ty area Bat Xat Nature Reserve is located in Bat Xat district of Lao Cai province, northwestern Vietnam. It was established in 2016 by the Decision No.1954/QD-UBND of the President of Lao Cai province “On the establishment of Bat Xat Nature Reserve” (DARD 2016) in order to conserve primeval forest ecosystems in the highlands, and in particular, the rare and endangered species of flora and fauna typical of Hoang Lien Son mountain region. -

Podophyllotoxin: History, Recent Advances and Future Prospects
biomolecules Review Podophyllotoxin: History, Recent Advances and Future Prospects Zinnia Shah 1 , Umar Farooq Gohar 1, Iffat Jamshed 1, Aamir Mushtaq 2 , Hamid Mukhtar 1 , Muhammad Zia-UI-Haq 3,*, Sebastian Ionut Toma 4,*, Rosana Manea 4,*, Marius Moga 4 and Bianca Popovici 4 1 Institute of Industrial Biotechnology (IIB), Government College University, Lahore 54000, Pakistan; [email protected] (Z.S.); [email protected] (U.F.G.); [email protected] (I.J.); [email protected] (H.M.) 2 Gulab Devi Institute of Pharmacy, Gulab Devi Educational Complex, Lahore 54000, Pakistan; [email protected] 3 Office of Research, Innovation & Commercialization, Lahore College for Women University, Lahore 54000, Pakistan 4 Faculty of Medicine, Transilvania University of Brasov, 500036 Brasov, Romania; [email protected] (M.M.); [email protected] (B.P.) * Correspondence: [email protected] (M.Z.-U.-H.); [email protected] (S.I.T.); [email protected] (R.M.) Abstract: Podophyllotoxin, along with its various derivatives and congeners are widely recognized as broad-spectrum pharmacologically active compounds. Etoposide, for instance, is the frontline chemotherapeutic drug used against various cancers due to its superior anticancer activity. It has recently been redeveloped for the purpose of treating cytokine storm in COVID-19 patients. Podophyllotoxin and its naturally occurring congeners have low bioavailability and almost all these initially discovered compounds cause systemic toxicity and development of drug resistance. Citation: Shah, Z.; Gohar, U.F.; Moreover, the production of synthetic derivatives that could suffice for the clinical limitations of Jamshed, I.; Mushtaq, A.; Mukhtar, these naturally occurring compounds is not economically feasible.