DATING PHYLOGENETICALLY BASAL EUDICOTS USING Rbcl SEQUENCES and MULTIPLE FOSSIL REFERENCE POINTS1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Plant List 2016
Established 1990 PLANT LIST 2016 European mail order website www.crug-farm.co.uk CRÛG FARM PLANTS • 2016 Welcome to our 2016 list hope we can tempt you with plenty of our old favourites as well as some exciting new plants that we have searched out on our travels. There has been little chance of us standing still with what has been going on here in 2015. The year started well with the birth of our sixth grandchild. January into February had Sue and I in Colombia for our first winter/early spring expedition. It was exhilarating, we were able to travel much further afield than we had previously, as the mountainous areas become safer to travel. We are looking forward to working ever closer with the Colombian institutes, such as the Medellin Botanic Gardens whom we met up with. Consequently we were absent from the RHS February Show at Vincent Square. We are finding it increasingly expensive participating in the London shows, while re-branding the RHS February Show as a potato event hardly encourages our type of customer base to visit. A long standing speaking engagement and a last minute change of date, meant that we missed going to Fota near Cork last spring, no such problem this coming year. We were pleasantly surprised at the level of interest at the Trgrehan Garden Rare Plant Fair, in Cornwall. Hopefully this will become an annual event for us, as well as the Cornwall Garden Society show in April. Poor Sue went through the wars having to have a rush hysterectomy in June, after some timely results revealed future risks. -

Coptis Trifolia Conservation Assessment
CONSERVATION ASSESSMENT for Coptis trifolia (L.) Salisb. Originally issued as Management Recommendations December 1998 Marty Stein Reconfigured-January 2005 Tracy L. Fuentes USDA Forest Service Region 6 and USDI Bureau of Land Management, Oregon and Washington CONSERVATION ASSESSMENT FOR COPTIS TRIFOLIA Table of Contents Page List of Tables ................................................................................................................................. 2 List of Figures ................................................................................................................................ 2 Summary........................................................................................................................................ 4 I. NATURAL HISTORY............................................................................................................. 6 A. Taxonomy and Nomenclature.......................................................................................... 6 B. Species Description ........................................................................................................... 6 1. Morphology ................................................................................................................... 6 2. Reproductive Biology.................................................................................................... 7 3. Ecological Roles ............................................................................................................. 7 C. Range and Sites -

Management of Cercospora Leaf Spot of Indian Spinach (Basella Alba L.) with BAU Bio-Fungicide and a Plant Growth Promoting Hormone
Universal Journal of Plant Science 4(4): 43-49, 2016 http://www.hrpub.org DOI: 10.13189/ujps.2016.040401 Management of Cercospora Leaf Spot of Indian Spinach (Basella alba L.) with BAU Bio-fungicide and a Plant Growth Promoting Hormone Md. Mohidul Hasan1,*, Nazia Binta Islam1, Shamima Naznin1, Md. Mobinul Islam1, 2 Kishowar-E-Mustarin 1Department of Plant Pathology, Hajee Mohammad Danesh Science and Technology University, Bangladesh 2Wheat Research Centre, Bangladesh Agricultural Research Institute Joydebpur, Bangladesh Copyright©2016 by authors, all rights reserved. Authors agree that this article remains permanently open access under the terms of the Creative Commons Attribution License 4.0 International License Abstract Trichoderma based BAU-biofungicide, Basellaceae is an indigenous, rapidly growing, tropical leafy chemical Carbendazim and a synthetic plant growth vegetable [1,2] commonly grown as backyard plant in the promoting (PGP) hormone have been used to study their home gardens. It is originated from India or Indonesia [3], effect on Cercospora leaf spot of Indian spinach. Number of however; it is also popular in tropical and subtropical region leaf, number of infected leaf, disease incidence, disease including Asia, America, Africa, Madagascar etc. [2]. The severity, area under disease progress curve (AUDPC), plant vegetable has other interesting common names in different height and plant weight were measured and significant region like Ceylon spinach, Malabar spinach, saan choy variations was found against different treatment -

Post Graduate Department of Chemistry Faculty of Physical and Material Sciences-2011 the University of Kashmir Hazratbal, Srinagar-190006
CORE Metadata, citation and similar papers at core.ac.uk Provided by Knowledge Repository Open Network Phytochemical Screening of Major Constituents of Various Folklore Medicinal Plants of Kashmir Valley DISSERTATION Submitted in partial fulfillment of the requirements provided for the award of the degree of Master of Philosophy In Chemistry By Sofi Mubashir Under the joint supervision of Dr. Syed Wajaht Amin Shah (Sr. Asstt. Professor, Deptt. of Chemistry, University of Kashmir) & Dr. Seema Akbar (Asstt. Director Chemistry, CCRUM, Srinagar) Post Graduate Department of Chemistry Faculty of Physical and Material Sciences-2011 The University of Kashmir Hazratbal, Srinagar-190006 Department of Chemistry University of Kashmir Srinagar-190006. CERTIFICATE This is to certify that Mr. Sofi Mubashir worked under our joint supervision for his M.Phil, studies “Phytochemical Screening of Major Constituents of Various Folklore Medicinal Plants of Kashmir Valley”. His work embodied in this dissertation is original. Mr. Sofi Mubashir has fulfilled all the formalities prior to submission of this dissertation. His work and conduct has been satisfactory. The dissertation is recommended for the award of M.Phil degree. (Co-Supervisor) (Supervisor) Dr. Seema Akbar Dr. Syed Wajaht Amin Shah Asstt. director (Chemistry) Sr. Asstt. Professor, CCRUM, Srinagar. Department of Chemistry, University of Kashmir. DEDICATION This study is dedicated to my parents who have always been there for me. Acknowledgement To begin with I feel highly thankful to my Supervisor Dr. Syed Wajaht Amin Shah and Co-supervisor Dr. Seema Akbar for their sustained and strenuous effort to enlarge my understanding of the topic at each step; for energizing me at every moment. -

Bio 308-Course Guide
COURSE GUIDE BIO 308 BIOGEOGRAPHY Course Team Dr. Kelechi L. Njoku (Course Developer/Writer) Professor A. Adebanjo (Programme Leader)- NOUN Abiodun E. Adams (Course Coordinator)-NOUN NATIONAL OPEN UNIVERSITY OF NIGERIA BIO 308 COURSE GUIDE National Open University of Nigeria Headquarters 14/16 Ahmadu Bello Way Victoria Island Lagos Abuja Office No. 5 Dar es Salaam Street Off Aminu Kano Crescent Wuse II, Abuja e-mail: [email protected] URL: www.nou.edu.ng Published by National Open University of Nigeria Printed 2013 ISBN: 978-058-434-X All Rights Reserved Printed by: ii BIO 308 COURSE GUIDE CONTENTS PAGE Introduction ……………………………………......................... iv What you will Learn from this Course …………………............ iv Course Aims ……………………………………………............ iv Course Objectives …………………………………………....... iv Working through this Course …………………………….......... v Course Materials ………………………………………….......... v Study Units ………………………………………………......... v Textbooks and References ………………………………........... vi Assessment ……………………………………………….......... vi End of Course Examination and Grading..................................... vi Course Marking Scheme................................................................ vii Presentation Schedule.................................................................... vii Tutor-Marked Assignment ……………………………….......... vii Tutors and Tutorials....................................................................... viii iii BIO 308 COURSE GUIDE INTRODUCTION BIO 308: Biogeography is a one-semester, 2 credit- hour course in Biology. It is a 300 level, second semester undergraduate course offered to students admitted in the School of Science and Technology, School of Education who are offering Biology or related programmes. The course guide tells you briefly what the course is all about, what course materials you will be using and how you can work your way through these materials. It gives you some guidance on your Tutor- Marked Assignments. There are Self-Assessment Exercises within the body of a unit and/or at the end of each unit. -

Bioavailability of Iron from Basella Alba and Amaranthus Hybridus Leaves Supplemented Diet in Iron Deficient Anaemic Albino Rats
BIOAVAILABILITY OF IRON FROM BASELLA ALBA AND AMARANTHUS HYBRIDUS LEAVES SUPPLEMENTED DIET IN IRON DEFICIENT ANAEMIC ALBINO RATS. BY Ceaser Antiya, MOSES DEPARTMENT OF BIOCHEMISTRY FACULTY OF SCIENCE AHMADU BELLO UNIVERSITY ZARIA JANUARY, 2016 i BIOAVAILABILITY OF IRON FROM BASELLA ALBA AND AMARANTHUS HYBRIDUS LEAVES SUPPLEMENTED DIET IN IRON DEFICIENT ANAEMIC ALBINO RATS. BY Ceaser Antiya MOSES, B.SC BIOCHEMISTRY (A.B.U) 2011 MSc/SCIE/44812/2012-2013 A DISSERTATION SUBMITTED TO THE SCHOOL OF POSTGRADUATE STUDIES, AHMADU BELLO UNIVERSITY, ZARIA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE AWARD OF MASTERS DEGREE IN BIOCHEMISTRY DEPARTMENT OF BIOCHEMISTRY FACULTY OF SCIENCE AHMADU BELLO UNIVERSITY, ZARIA NIGERIA JANUARY, 2016 ii DECLARATION I hereby declare that the work in this dissertation entitled ―BIOAVAILABILITY OF IRON FROM BASELLA ALBA AND AMARANTHUS HYBRIDUS LEAVES SUPPLEMENTED DIET IN IRON DEFICIENT ANAEMIC ALBINO RATS” has been performed by me in the Department of Biochemistry under the supervision of Mr. O. A. Owolabi and Prof. E. Oyinke. The information herein derived from literature has been duly acknowledged in the text and a list of references provided. No part of this dissertationwas previously presented for another degree or diploma at any university to the best of my knowledge. …………………………………. ……………………… …………………… Moses Ceaser Antiya Signature Date iii CERTIFICATION This dissertation entitled ―BIOAVAILABILITY OF IRON FROM BASELLA ALBA AND AMARANTHUS HYBRIDUS LEAVES SUPPLEMENTED DIET IN IRON DEFICIENT ANAEMIC ALBINO RATS” by Ceaser, Antiya MOSES meets the regulations governing the award of the degree of MASTER of Science Ahmadu Bello University, and is approved for its contribution to knowledge and literary presentation. -

Dry Forest Trees of Madagascar
The Red List of Dry Forest Trees of Madagascar Emily Beech, Malin Rivers, Sylvie Andriambololonera, Faranirina Lantoarisoa, Helene Ralimanana, Solofo Rakotoarisoa, Aro Vonjy Ramarosandratana, Megan Barstow, Katharine Davies, Ryan Hills, Kate Marfleet & Vololoniaina Jeannoda Published by Botanic Gardens Conservation International Descanso House, 199 Kew Road, Richmond, Surrey, TW9 3BW, UK. © 2020 Botanic Gardens Conservation International ISBN-10: 978-1-905164-75-2 ISBN-13: 978-1-905164-75-2 Reproduction of any part of the publication for educational, conservation and other non-profit purposes is authorized without prior permission from the copyright holder, provided that the source is fully acknowledged. Reproduction for resale or other commercial purposes is prohibited without prior written permission from the copyright holder. Recommended citation: Beech, E., Rivers, M., Andriambololonera, S., Lantoarisoa, F., Ralimanana, H., Rakotoarisoa, S., Ramarosandratana, A.V., Barstow, M., Davies, K., Hills, BOTANIC GARDENS CONSERVATION INTERNATIONAL (BGCI) R., Marfleet, K. and Jeannoda, V. (2020). Red List of is the world’s largest plant conservation network, comprising more than Dry Forest Trees of Madagascar. BGCI. Richmond, UK. 500 botanic gardens in over 100 countries, and provides the secretariat to AUTHORS the IUCN/SSC Global Tree Specialist Group. BGCI was established in 1987 Sylvie Andriambololonera and and is a registered charity with offices in the UK, US, China and Kenya. Faranirina Lantoarisoa: Missouri Botanical Garden Madagascar Program Helene Ralimanana and Solofo Rakotoarisoa: Kew Madagascar Conservation Centre Aro Vonjy Ramarosandratana: University of Antananarivo (Plant Biology and Ecology Department) THE IUCN/SSC GLOBAL TREE SPECIALIST GROUP (GTSG) forms part of the Species Survival Commission’s network of over 7,000 Emily Beech, Megan Barstow, Katharine Davies, Ryan Hills, Kate Marfleet and Malin Rivers: BGCI volunteers working to stop the loss of plants, animals and their habitats. -

The First Comprehensive Phylogeny of Coptis (Ranunculaceae) and Its Implications for Character Evolution and Classification
RESEARCH ARTICLE The First Comprehensive Phylogeny of Coptis (Ranunculaceae) and Its Implications for Character Evolution and Classification Kun-Li Xiang1,2, Sheng-Dan Wu3, Sheng-Xian Yu1, Yang Liu4, Florian Jabbour5, Andrey S. Erst6,7, Liang Zhao8, Wei Wang1*, Zhi-Duan Chen1 1 State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, 100093, China, 2 University of Chinese Academy of Sciences, Beijing, 100049, China, 3 College of Life Sciences, Shanxi Normal University, Linfen, 041004, China, 4 Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, Connecticut, 06269–3043, United States of America, 5 Muséum national d’Histoire naturelle, Institut de Systématique, Evolution, Biodiversité, UMR 7205 ISYEB MNHN/CNRS/UPMC/EPHE, Sorbonne Universités, Paris, 75005, France, 6 Central Siberian Botanical Garden of the Siberian Branch of Russian Academy of Sciences, Zolotodolinskaya str. 101, Novosibirsk, 630090, Russia, 7 Laboratory of Systematics and Phylogeny of Plants, National Research Tomsk State University, prospekt Lenina 36, Tomsk, 634050, Russia, 8 College of Life Sciences, Northwest A&F University, Yangling, Shaanxi, 712100, China * [email protected] OPEN ACCESS Citation: Xiang K-L, Wu S-D, Yu S-X, Liu Y, Jabbour F, Erst AS, et al. (2016) The First Comprehensive Abstract Phylogeny of Coptis (Ranunculaceae) and Its Implications for Character Evolution and Coptis (Ranunculaceae) contains 15 species and is one of the pharmaceutically most Classification. PLoS ONE 11(4): e0153127. important plant genera in eastern Asia. Understanding of the evolution of morphological doi:10.1371/journal.pone.0153127 characters and phylogenetic relationships within the genus is very limited. -

Written Report on a Joint AGS/Merlin Trust Funded Trip Charlotte Reynolds Merlin 607 30Th November – 16Th December 2013
Alpine Adventures in Argentina: In search of Violas Written report on a Joint AGS/Merlin Trust Funded Trip Charlotte Reynolds Merlin 607 30th November – 16th December 2013 Charlotte Reynolds 1 Contents Introduction - page 3 Day by day log - page 4 Monday 2nd December – Laguna Blanca - page 4 Tuesday 3rd December – Laguna del Burro & Rahue Pass - page 6 Wednesday 4th December – Primeros Pinos - page 8 Thursday 5th December - Volcan Batea Mahuida - page 9 Friday 6th December - Rio Litran - page 11 Saturday 7th December – Copahue - page 12 Sunday 8th December – Salta del Agrio & Cascada del Agrio - page 13 Monday 9th December - Cordon del Cajon Chico - page 14 Tuesday 10th December - Chos Malal - page 15 Wednesday 11th December – Tromen National Park - page 15 Thursday 12th December - Cerro Wayle -page 16 Friday 13th December - Lagunas Epu Lauquen - page 17 Plant family index – page 19 Violas - page 19 Trees and shrubs – page 23 Cushions - page 33 Bulbs - page 38 Orchids - page 42 Cactus – page 43 Wetland - page 45 Perennials – page 50 Conclusion – page 64 Charlotte Reynolds 2 Introduction & Background The Alpine Garden Society ran this tour to Northern Patagonia led by Martin Sheader, an expert in flora of the area. Patagonia is a region in the far south of South America which is divided between Chile and Argentina. We toured the Argentine Patagonian province Neuquen travelling around Zapala, Villa Pehuenia, Caviahue and Chos Malal. The landscape in these areas was dry, grassland steppes in the main. However we also saw a wide-range of micro-habitats, due to the different localised conditions. For instance, boggy wetlands caused by snow melt, shady cliff faces, Auracaria forest floors, rocky crevices and pumice-covered cliffs to name a few. -
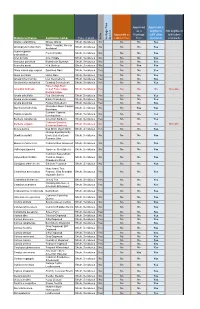
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Calibrated Chronograms, Fossils, Outgroup Relationships, and Root Priors: Re-Examining the Historical Biogeography of Geraniales
bs_bs_banner Biological Journal of the Linnean Society, 2014, 113, 29–49. With 4 figures Calibrated chronograms, fossils, outgroup relationships, and root priors: re-examining the historical biogeography of Geraniales KENNETH J. SYTSMA1,*, DANIEL SPALINK1 and BRENT BERGER2 1Department of Botany, University of Wisconsin, Madison, WI 53706, USA 2Department of Biological Sciences, St. John’s University, Queens, NY 11439, USA Received 26 November 2013; revised 23 February 2014; accepted for publication 24 February 2014 We re-examined the recent study by Palazzesi et al., (2012) published in the Biological Journal of the Linnean Society (107: 67–85), that presented the historical diversification of Geraniales using BEAST analysis of the plastid spacer trnL–F and of the non-coding nuclear ribosomal internal transcribed spacers (ITS). Their study presented a set of new fossils within the order, generated a chronogram for Geraniales and other rosid orders using fossil-based priors on five nodes, demonstrated an Eocene radiation of Geraniales (and other rosid orders), and argued for more recent (Pliocene–Pleistocene) and climate-linked diversification of genera in the five recognized families relative to previous studies. As a result of very young ages for the crown of Geraniales and other rosid orders, unusual relationships of Geraniales to other rosids, and apparent nucleotide substitution saturation of the two gene regions, we conducted a broad series of BEAST analyses that incorporated additional rosid fossil priors, used more accepted rosid ordinal -

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m.