HSH2D Contributes to Methotrexate Resistance in Human T‑Cell Acute Lymphoblastic Leukaemia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Molecular Effects of Fermentation in the Gut and Its Relevance for Metabolism and Satiety
Molecular effects of fermentation in the gut and its relevance for metabolism and satiety Daniëlle Haenen Thesis committee Promotor Prof. Dr Michael R. Müller Professor of Nutrition, Metabolism and Genomics Wageningen University Co-promotors Dr Guido J.E.J. Hooiveld Assistant professor, Division of Human Nutrition Wageningen University Prof. Dr Bas Kemp Professor of Adaptation Physiology Wageningen University Other members Prof. Dr Renger F. Witkamp, Wageningen University Prof. Dr Wouter H. Hendriks, Wageningen University Prof. Dr Michiel Kleerebezem, Wageningen University Prof. Dr Albert K. Groen, University of Groningen This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of fermentation in the gut and its relevance for metabolism and satiety Daniëlle Haenen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Wednesday 18 September 2013 at 1.30 p.m. in the Aula. Daniëlle Haenen Molecular effects of fermentation in the gut and its relevance for metabolism and satiety 210 pages PhD thesis, Wageningen University, Wageningen, NL (2013) With references, with summary in Dutch ISBN 978-94-6173-667-3 Abstract Dietary fibres, the edible parts of plants that are resistant to digestion and absorption in the human small intestine, were shown to be important in the prevention of obesity and the metabolic syndrome. This association can partially be attributed to a fibre-induced increase in satiety. -

Mapping and Characterization of the Interaction Network of ALK Receptor Tyrosine Kinase Using the Mammalian Membrane Two-Hybrid (Mamth) Assay
Mapping and Characterization of the Interaction Network of ALK Receptor Tyrosine Kinase using the Mammalian Membrane Two-Hybrid (MaMTH) Assay by Farzaneh Aboualizadeh A thesis submitted in conformity with the requirements for the degree of Master of Science Graduate Department of Biochemistry University of Toronto © Copyright by Farzaneh Aboualizadeh 2018 Mapping and Characterization of the Interaction Network of ALK Receptor Tyrosine Kinase using the Mammalian Membrane Two-Hybrid (MaMTH) Assay Farzaneh Aboualizadeh Master of Science Graduate Department of Biochemistry University of Toronto 2018 Abstract Anaplastic Lymphoma Kinase (ALK) which belongs to the Receptor Tyrosine Kinases (RTKs) play a critical role in development and progression of many type of cancers. To design new treatments for ALK associated human diseases, it is essential to study the interactions of the protein to better understand the biological significance of its protein-protein interactions (PPIs) and how they contribute to ALK regulation and function. Novel high-throughput technology, the Mammalian Membrane Two-Hybrid (MaMTH), was used in this study to generate a targeted protein-protein interaction map (interactome) of full length ALK. Biased screening of 150 predicted and previously known ALK interactors were performed and 34 identified as novel ALK interactors. The active mutant cells stably expressing ALK F1245C, F1245I and L1196M have been generated which showed more phosphorylation of ALK (pALK1604) as compared to wild type. Moreover, these active mutants exhibited more phosphorylation of downstream signaling ERK1/2 as compared to wild type. ii Acknowledgments I would like to express my sincere gratitude to my supervisor, Dr. Igor Stagljar, for the useful advices, remarks and engagement through the learning process of this master thesis. -

Genome-Wide Screening Identifies Genes and Biological Processes
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 10-12-2018 Genome-Wide Screening Identifies Genes and Biological Processes Implicated in Chemoresistance and Oncogene-Induced Apoptosis Tengyu Ko Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Part of the Cancer Biology Commons, Cell Biology Commons, and the Genomics Commons Recommended Citation Ko, Tengyu, "Genome-Wide Screening Identifies Genes and Biological Processes Implicated in Chemoresistance and Oncogene- Induced Apoptosis" (2018). LSU Doctoral Dissertations. 4715. https://digitalcommons.lsu.edu/gradschool_dissertations/4715 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. GENOME-WIDE SCREENING IDENTIFIES GENES AND BIOLOGICAL PROCESSES IMPLICATED IN CHEMORESISTANCE AND ONCOGENE- INDUCED APOPTOSIS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical and Veterinary Medical Sciences through the Department of Comparative Biomedical Sciences by Tengyu Ko B.S., University of California, Santa Barbara 2010 December 2018 ACKNOWLEDGEMENTS I would like to express my sincerest gratitude to my major supervisor Dr. Shisheng Li for giving me the opportunity to join his team and the freedom to pursue projects. I appreciate all of his thoughts and efforts. Truly, none of these findings would be possible without his supervisions, supports, insightful discussions, and patience. -

(A) Sample Contribution to the PLS Model
SUPPLEMENTARY DATA Supplementary Figure 1. Correlation of levels of auto-antibodies with disease duration. (A) Sample contribution to the PLS model. The bars represent the distances of individual samples to the PLS model, which represent the contribution of individual samples to the separation between T1DM and T2DM/NGT captured by the PLS model. A small distance indicates high contribution of the corresponding sample to the separation captured by the PLS model. The numbers in parenthesis in the x- axis labels of T1DM samples represent the disease durations in months. (B) Scatterplot of disease duration and sample contribution to the separation between T1DM and T2DM/NGT captured by the PLS model (i.e. distances of samples to the PLS model). The distances showed no significant correlation (r = –0.465 and P = 0.070) with disease durations of the individual samples. ©2014 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-1566/-/DC1 SUPPLEMENTARY DATA Supplementary Figure 2. Expression of EEF1A1 and UBE2L3 in human tissues. Immunohistochemistry of tissue array showed that (A) EEF1A1 and (B) UBE2L3 were expressed in skin, liver, kidney, stomach, intestine, lung, adrenal gland, and thyroid gland as well as pancreas. Original magnification 400. 헑 ©2014 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-1566/-/DC1 SUPPLEMENTARY DATA Supplementary Figure 3. Detection of EEF1A1 and UBE2L3 auto-antibodies by immunoblotting. Three pairs of T1DM patient samples were selected from the second cohort based on their ELISA absorbance values categorized in three groups: high: >1.3, medium: <1.3 and >0.5, and low: <0.5 absorbance at 450 nm (A450nm). -
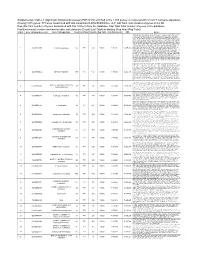
(FDR<0.05) Enriched in the 1015 Genes Co-Clustered with Known T
Supplementary Table 3. Significant biological processes (FDR<0.05) enriched in the 1,015 genes co-clustered with known T cell gene signatures. Among 1,015 genes, 771 were associated with GO annotation in DAVID database v6.7. List Total: total number of genes in my list. Pop Hits: total number of genes associated with this GO term from the database. Pop Total: total number of genes in the database. Fold Enrichment: relative enrichment ratio, calculated by (Count)/(List Total) divided by (Pop Hits)/(Pop Total). Index Gene Ontology Accession Gene Ontology Name Count List Total Pop Hits Pop Total Fold Enrichment FDR Genes AQP9, C1QC, B2M, LILRA1, LILRA2, CLEC4E, S1PR4, LILRA4, IFNG, LILRA6, CLEC4A, VNN1, ERAP2, FAS, CRTAM, C5AR1, GBP5, NCF2, NCF1, NCF4, SERPING1, HLA-DQA2, HLA-DQA1, PDCD1LG2, LILRB1, CCR9, C1QA, C1QB, LILRB2, CCR7, CCR6, UNC13D, CCR5, CD40LG, CCR4, LILRB3, CCR2, LILRB4, HLA-DPA1, VSIG4, HLA-DRA, IL1R2, IL1R1, HLA-DRB1, OAS3, ACP5, OAS1, OAS2, CD74, IFI35, ZAP70, FCER1G, HLA-DRB5, HLA-DPB1, HLA-DOA, HLA-DOB, DHX58, BLNK, IL23R, KIR2DS4, CD300C, SLAMF7, OASL, RGS1, APOL1, CD300A, HMHB1, CD209, CLEC7A, LY86, LY9, CLNK, FCRL4, SH2D1A, NOD2, HAMP, CCL3L1, CCL3L3, TICAM2, ICAM1, GZMA, CMKLR1, LY96, WAS, IL18BP, LAX1, TNFSF12- TNFSF13, HLA-DQB1, CSF2, GPR183, CCR1, GPR65, CXCL9, NCF1C, IL7R, CLEC10A, CCL24, CCL22, CYP27B1, CCL23, FCGR1C, FTHL3, FCGR1A, FCGR1B, BCL3, C2, CD27, CD28, FYB, IL18R1, IL7, CD1C, CTLA4, CCL19, CD1B, CD1A, TRIM22, CD180, CD1E, CCL18, CCL17, CCL13, FCGR2B, FCGR2C, P2RY14, LIME1, CD14, IL16, IL18, TLR1, TNFSF15, -

The HIV-1 Gp120/V3 Modifies the Response of Uninfected CD4 T Cells
Morou et al. Journal of Translational Medicine 2011, 9:160 http://www.translational-medicine.com/content/9/1/160 RESEARCH Open Access The HIV-1 gp120/V3 modifies the response of uninfected CD4 T cells to antigen presentation: mapping of the specific transcriptional signature Antigone K Morou1, Filippos Porichis2, Elias Krambovitis3, George Sourvinos1, Demetrios A Spandidos1 and Alexandros Zafiropoulos4* Abstract Background: The asymptomatic phase of HIV-1 infection is characterized by a progressive depletion of uninfected peripheral effector/memory CD4+ T cells that subsequently leads to immune dysfunction and AIDS symptoms. We have previously demonstrated that the presence of specific gp120/V3 peptides during antigen presentation can modify the activation of normal T-cells leading to altered immune function. The aim of the present study was to map the specific transcriptional profile invoked by an HIV-1/V3 epitope in uninfected T cells during antigen presentation. Methods: We exposed primary human peripheral blood monocytes to V3 lipopeptides using a liposome delivery system followed by a superantigen-mediated antigen presentation system. We then evaluated the changes in the T-cell transcriptional profile using oligonucleotide microarrays and performed Ingenuity Pathway Analysis (IPA) and DAVID analysis. The results were validated using realtime PCR, FACS, Western blotting and immunofluorescence. Results: Our results revealed that the most highly modulated transcripts could almost entirely be categorized as related to the cell cycle or transcriptional regulation. The most statistically significant enriched categories and networks identified by IPA were associated with cell cycle, gene expression, immune response, infection mechanisms, cellular growth, proliferation and antigen presentation. Canonical pathways involved in energy and cell cycle regulation, and in the co-activation of T cells were also enriched. -

Current Challenges Towards the Development of a Blood Test for Parkinson’S Disease
Diagnostics 2014, 4, 153-164; doi:10.3390/diagnostics4040153 OPEN ACCESS diagnostics ISSN 2075-4418 www.mdpi.com/journal/diagnostics/ Review Current Challenges Towards the Development of a Blood Test for Parkinson’s Disease Jose A. Santiago and Judith A. Potashkin * The Cellular and Molecular Pharmacology Department, The Chicago Medical School, Rosalind Franklin University of Medicine and Science, North Chicago, IL 60064-3037, USA; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-847-578-8677; Fax: +1-847-578-3268. External Editor: Ludmilla A. Morozova-Roche Received: 9 September 2014; in revised form: 8 October 2014 / Accepted: 11 October 2014 / Published: 22 October 2014 Abstract: Parkinson’ disease (PD) is the second most prevalent neurodegenerative disease worldwide. To date, there is no disease-modifying agent, and current medical treatment only provides symptomatic benefits. Early diagnosis of PD would be useful in clinical practice to identify patients for clinical trials, test potential drugs and neuroprotective agents and track their therapeutic effect. Considerable progress has been made in the discovery and validation of diagnostic biomarkers for PD. In particular, blood-based biomarkers have shown promise in identifying PD patients in samples from independent clinical trials. Evaluation of these biomarkers in de novo patients and individuals at risk for PD remains a top priority. Here, we review the current advances and challenges toward the clinical translation of these biomarkers into a blood-based test for PD. Keywords: biomarkers; blood; Parkinson’s disease 1. Introduction Parkinson’s disease (PD) is a devastating neurodegenerative disease characterized by the progressive deterioration of the dopaminergic system in the substantia nigra pars compacta (SNpc). -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -

(DARPP-32) and Survival in Breast Cancer: a Retrospective Analysis of Protein and Mrna Expression Shreeya Kotecha1, Marie N
www.nature.com/scientificreports OPEN Dopamine and cAMP-regulated phosphoprotein 32 kDa (DARPP-32) and survival in breast cancer: a retrospective analysis of protein and mRNA expression Shreeya Kotecha1, Marie N. Lebot1, Bhudsaban Sukkarn1, Graham Ball 2, Paul M. Moseley1, Stephen Y. Chan1, Andrew R. Green1, Emad Rakha1, Ian O. Ellis1, Stewart G. Martin1 & Sarah J. Storr 1* Dopamine and cAMP regulated phosphoprotein 32 kDa (DARPP-32) also known as phosphoprotein phosphatase-1 regulatory subunit 1B and encoded by the PPP1R1B gene is an inhibitor of protein phosphatase-1 and protein kinase A. DARPP-32 is expressed in a wide range of epithelial cells and some solid tumours; however, its role in breast cancer is only partially defned. DARPP-32 expression was determined using immunohistochemistry in two independent cohorts of early stage invasive breast cancer patients (discovery n = 1352; validation n = 1655), and 112 HER2 positive breast cancer patients treated with trastuzumab and adjuvant chemotherapy. PPP1R1B mRNA expression was assessed in the METABRIC cohort (n = 1980), using artifcial neural network analysis to identify associated genes. In the discovery cohort, low nuclear expression of DARPP-32 was signifcantly associated with shorter survival (P = 0.041), which was independent of other prognostic variables (P = 0.019). In the validation cohort, low cytoplasmic and nuclear expression was signifcantly associated with shorter survival (both P = 0.002), with cytoplasmic expression independent of other prognostic variables (P = 0.023). Stronger associations with survival in oestrogen receptor (ER) positive disease were observed. In patients treated with trastuzumab, low nuclear expression was signifcantly associated with adverse progression-free survival (P = 0.031). -

Water-Pipe Smoking Exposure Deregulates a Set of Genes Associated with Human Head and Neck Cancer Development and Prognosis
toxics Article Water-Pipe Smoking Exposure Deregulates a Set of Genes Associated with Human Head and Neck Cancer Development and Prognosis 1, 2,3, 1 1 Vanessa M. López-Ozuna y , Ishita Gupta y, Ryan Liu Chen Kiow , Emad Matanes , Hadeel Kheraldine 2,3 , Amber Yasmeen 1, Ashraf Khalil 4, Semir Vranic 2 , Ala-Eddin Al Moustafa 2,3 and Halema F Al Farsi 2,* 1 Segal Cancer Center, Lady Davis Institute of Medical Research, JGH, McGill University, Montreal, QC H3T IE2, Canada; [email protected] (V.M.L.-O.); [email protected] (R.L.C.K.); [email protected] (E.M.); [email protected] (A.Y.) 2 College of Medicine, QU Health, Qatar University, Doha PO Box 2713, Qatar; [email protected] (I.G.); [email protected] (H.K.); [email protected] (S.V.); [email protected] (A.-E.A.M.) 3 Biomedical Research Centre, Qatar University, Doha PO Box 2713, Qatar 4 College of Pharmacy, QU Health, Qatar University, Doha PO Box 2713, Qatar; [email protected] * Correspondence: [email protected]; Tel.: +1-(974)-4403-7840 Both authors contributed equally to this work. y Received: 30 July 2020; Accepted: 14 September 2020; Published: 18 September 2020 Abstract: Water-pipe smoking (WPS) is becoming the most popular form of tobacco use among the youth, especially in the Middle East, replacing cigarettes rapidly and becoming a major risk of tobacco addiction worldwide. Smoke from WPS contains similar toxins as those present in cigarette smoke and is linked directly with different types of cancers including lung and head and neck (HN) carcinomas.