Increased Copy Number Variation of Mtdna in an Array-Based Digital
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dual Role of the Mitochondrial Protein Frataxin in Astrocytic Tumors
Laboratory Investigation (2011) 91, 1766–1776 & 2011 USCAP, Inc All rights reserved 0023-6837/11 $32.00 Dual role of the mitochondrial protein frataxin in astrocytic tumors Elmar Kirches1, Nadine Andrae1, Aline Hoefer2, Barbara Kehler1,3, Kim Zarse4, Martin Leverkus3, Gerburg Keilhoff5, Peter Schonfeld5, Thomas Schneider6, Annette Wilisch-Neumann1 and Christian Mawrin1 The mitochondrial protein frataxin (FXN) is known to be involved in mitochondrial iron homeostasis and iron–sulfur cluster biogenesis. It is discussed to modulate function of the electron transport chain and production of reactive oxygen species (ROS). FXN loss in neurons and heart muscle cells causes an autosomal-dominant mitochondrial disorder, Friedreich’s ataxia. Recently, tumor induction after targeted FXN deletion in liver and reversal of the tumorigenic phenotype of colonic carcinoma cells following FXN overexpression were described in the literature, suggesting a tumor suppressor function. We hypothesized that a partial reversal of the malignant phenotype of glioma cells should occur after FXN transfection, if the mitochondrial protein has tumor suppressor functions in these brain tumors. In astrocytic brain tumors and tumor cell lines, we observed reduced FXN levels compared with non-neoplastic astrocytes. Mitochondrial content (citrate synthase activity) was not significantly altered in U87MG glioblastoma cells stably overexpressing FXN (U87-FXN). Surprisingly, U87-FXN cells exhibited increased cytoplasmic ROS levels, although mitochondrial ROS release was attenuated by FXN, as expected. Higher cytoplasmic ROS levels corresponded to reduced activities of glutathione peroxidase and catalase, and lower glutathione content. The defect of antioxidative capacity resulted in increased susceptibility of U87-FXN cells against oxidative stress induced by H2O2 or buthionine sulfoximine. -

The Pathogenetic Role of Β-Cell Mitochondria in Type 2 Diabetes
236 3 Journal of M Fex et al. Mitochondria in β-cells 236:3 R145–R159 Endocrinology REVIEW The pathogenetic role of β-cell mitochondria in type 2 diabetes Malin Fex1, Lisa M Nicholas1, Neelanjan Vishnu1, Anya Medina1, Vladimir V Sharoyko1, David G Nicholls1, Peter Spégel1,2 and Hindrik Mulder1 1Department of Clinical Sciences in Malmö, Unit of Molecular Metabolism, Lund University Diabetes Centre, Clinical Research Center, Malmö University Hospital, Lund University, Malmö, Sweden 2Department of Chemistry, Center for Analysis and Synthesis, Lund University, Sweden Correspondence should be addressed to H Mulder: [email protected] Abstract Mitochondrial metabolism is a major determinant of insulin secretion from pancreatic Key Words β-cells. Type 2 diabetes evolves when β-cells fail to release appropriate amounts f TCA cycle of insulin in response to glucose. This results in hyperglycemia and metabolic f coupling signal dysregulation. Evidence has recently been mounting that mitochondrial dysfunction f oxidative phosphorylation plays an important role in these processes. Monogenic dysfunction of mitochondria is a f mitochondrial transcription rare condition but causes a type 2 diabetes-like syndrome owing to β-cell failure. Here, f genetic variation we describe novel advances in research on mitochondrial dysfunction in the β-cell in type 2 diabetes, with a focus on human studies. Relevant studies in animal and cell models of the disease are described. Transcriptional and translational regulation in mitochondria are particularly emphasized. The role of metabolic enzymes and pathways and their impact on β-cell function in type 2 diabetes pathophysiology are discussed. The role of genetic variation in mitochondrial function leading to type 2 diabetes is highlighted. -

Src Inhibitors Modulate Frataxin Protein Levels
Human Molecular Genetics, 2015, Vol. 24, No. 15 4296–4305 doi: 10.1093/hmg/ddv162 Advance Access Publication Date: 6 May 2015 Original Article ORIGINAL ARTICLE Src inhibitors modulate frataxin protein levels Downloaded from https://academic.oup.com/hmg/article/24/15/4296/2453025 by guest on 28 September 2021 Fabio Cherubini1, Dario Serio1, Ilaria Guccini1, Silvia Fortuni1,2, Gaetano Arcuri1, Ivano Condò1, Alessandra Rufini1,2, Shadman Moiz1, Serena Camerini3, Marco Crescenzi3, Roberto Testi1,2 and Florence Malisan1,* 1Laboratory of Signal Transduction, Department of Biomedicine and Prevention, University of Rome ‘Tor Vergata’, Via Montpellier 1, 00133 Rome, Italy, 2Fratagene Therapeutics Ltd, 22 Northumberland Rd, Dublin, Ireland and 3Department of Cell Biology and Neurosciences, Italian National Institute of Health, Viale Regina Elena, 299, 00161 Rome, Italy *To whom correspondence should be addressed at: Laboratory of Signal Transduction, Department of Biomedicine and Prevention, University of Rome ‘Tor Vergata’, Via Montpellier 1, 00133 Rome, Italy. Tel: +39 0672596501; Fax: +39 0672596505; Email: [email protected] Abstract Defective expression of frataxin is responsible for the inherited, progressive degenerative disease Friedreich’s Ataxia (FRDA). There is currently no effective approved treatment for FRDA and patients die prematurely. Defective frataxin expression causes critical metabolic changes, including redox imbalance and ATP deficiency. As these alterations are known to regulate the tyrosine kinase Src, we investigated whether Src might in turn affect frataxin expression. We found that frataxin can be phosphorylated by Src. Phosphorylation occurs primarily on Y118 and promotes frataxin ubiquitination, a signal for degradation. Accordingly, Src inhibitors induce accumulation of frataxin but are ineffective on a non-phosphorylatable frataxin- Y118F mutant. -
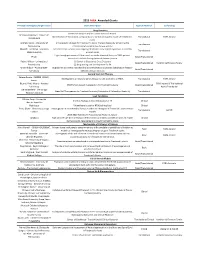
Grantsummaries 2014-2017 Jf
2015 FARA Awarded Grants Principal Investigator/Organization Grant Description Type of Research Co-funding Drug Discovery 2014 Keith Michael Andrus Cardiac Research Award: Veronique Monnier - Université Identification of therapeutic compounds on a cardiac Drosophila model of Friedreich’s Translational FARA Ireland Paris Diderot ataxia Andrew Dancis - University of A therapeutic strategy for Friedreich's ataxia: Frataxin bypass by enhancing the Translational Pennsylvania mitochondrial cysteine desulfurase activity Edward L. Grabczyk - Louisiana Small molecule induced exon skipping of MLH3 to slow repeat expansion in an FRDA Translational State University mousel model High throughput screen of Pfizer small molecule chemical library in FRDA patient- Pfizer Basic/Translational derived cells to look for regulators of frataxin protein Robert Wilson - University of (1) Center of Excellence: Drug Discovery Basic/Translational Hamilton & Finneran Family Pennsylvania (2) Drug testing and development for FA Omar Khdour - Arizona State Approaches to correct mitochondrial abnormalities and synaptic pathology in frataxin- Basic/Translational University deficient sensory neurons Gene & Stem Cell Therapy Helene Puccio - INSERM, IGBMC, Development of neuronal gene therapy for the treatment of FRDA Translational FARA Ireland France Ricardo Pinto Mouro - Harvard FARA Ireland & The National CRISPR/Cas9-based Therapeutics for Friedreich's ataxia Basic/Translational University Ataxia Foundation Joel Gottesfeld - The Scripps Stem Cell Therapeutics for Friedreich's -

Oxidative Stress, Mitochondrial Dysfunction, and Neuroprotection of Polyphenols with Respect to Resveratrol in Parkinson’S Disease
biomedicines Review Oxidative Stress, Mitochondrial Dysfunction, and Neuroprotection of Polyphenols with Respect to Resveratrol in Parkinson’s Disease Heng-Chung Kung 1,2 , Kai-Jung Lin 1,3 , Chia-Te Kung 4,* and Tsu-Kung Lin 1,5,6,* 1 Center for Mitochondrial Research and Medicine, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung 83301, Taiwan; [email protected] (H.-C.K.); [email protected] (K.-J.L.) 2 Department of Biology, Krieger School of Arts and Sciences, Johns Hopkins University, Baltimore, MD 21218, USA 3 Department of Family Medicine, National Taiwan University Hospital, Taipei 100225, Taiwan 4 Department of Emergency Medicine, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung 83301, Taiwan 5 Department of Neurology, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung 83301, Taiwan 6 Center of Parkinson’s Disease, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung 83301, Taiwan * Correspondence: [email protected] (C.-T.K.); [email protected] (T.-K.L.) Abstract: Parkinson’s disease (PD) is the second most common neurodegenerative disease and is characterized by dopaminergic neuronal loss. The exact pathogenesis of PD is complex and not yet completely understood, but research has established the critical role mitochondrial dysfunction plays in the development of PD. As the main producer of cytosolic reactive oxygen species (ROS), mito- chondria are particularly susceptible to oxidative stress once an imbalance between ROS generation and the organelle’s antioxidative system occurs. An overabundance of ROS in the mitochondria can Citation: Kung, H.-C.; Lin, K.-J.; lead to mitochondrial dysfunction and further vicious cycles. -

Strand Breaks for P53 Exon 6 and 8 Among Different Time Course of Folate Depletion Or Repletion in the Rectosigmoid Mucosa
SUPPLEMENTAL FIGURE COLON p53 EXONIC STRAND BREAKS DURING FOLATE DEPLETION-REPLETION INTERVENTION Supplemental Figure Legend Strand breaks for p53 exon 6 and 8 among different time course of folate depletion or repletion in the rectosigmoid mucosa. The input of DNA was controlled by GAPDH. The data is shown as ΔCt after normalized to GAPDH. The higher ΔCt the more strand breaks. The P value is shown in the figure. SUPPLEMENT S1 Genes that were significantly UPREGULATED after folate intervention (by unadjusted paired t-test), list is sorted by P value Gene Symbol Nucleotide P VALUE Description OLFM4 NM_006418 0.0000 Homo sapiens differentially expressed in hematopoietic lineages (GW112) mRNA. FMR1NB NM_152578 0.0000 Homo sapiens hypothetical protein FLJ25736 (FLJ25736) mRNA. IFI6 NM_002038 0.0001 Homo sapiens interferon alpha-inducible protein (clone IFI-6-16) (G1P3) transcript variant 1 mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 GALNTL5 NM_145292 0.0001 (GALNT15) mRNA. STIM2 NM_020860 0.0001 Homo sapiens stromal interaction molecule 2 (STIM2) mRNA. ZNF645 NM_152577 0.0002 Homo sapiens hypothetical protein FLJ25735 (FLJ25735) mRNA. ATP12A NM_001676 0.0002 Homo sapiens ATPase H+/K+ transporting nongastric alpha polypeptide (ATP12A) mRNA. U1SNRNPBP NM_007020 0.0003 Homo sapiens U1-snRNP binding protein homolog (U1SNRNPBP) transcript variant 1 mRNA. RNF125 NM_017831 0.0004 Homo sapiens ring finger protein 125 (RNF125) mRNA. FMNL1 NM_005892 0.0004 Homo sapiens formin-like (FMNL) mRNA. ISG15 NM_005101 0.0005 Homo sapiens interferon alpha-inducible protein (clone IFI-15K) (G1P2) mRNA. SLC6A14 NM_007231 0.0005 Homo sapiens solute carrier family 6 (neurotransmitter transporter) member 14 (SLC6A14) mRNA. -

Stríbrná and Julius Lukes Hassan Hashimi, Lindsay Mcdonald, Eva
Bioenergetics: Trypanosome Letm1 Protein Is Essential for Mitochondrial Potassium Homeostasis Hassan Hashimi, Lindsay McDonald, Eva Stríbrná and Julius Lukes J. Biol. Chem. 2013, 288:26914-26925. doi: 10.1074/jbc.M113.495119 originally published online July 26, 2013 Access the most updated version of this article at doi: 10.1074/jbc.M113.495119 Find articles, minireviews, Reflections and Classics on similar topics on the JBC Affinity Sites. Alerts: • When this article is cited • When a correction for this article is posted Click here to choose from all of JBC's e-mail alerts Supplemental material: http://www.jbc.org/content/suppl/2013/07/26/M113.495119.DC1.html This article cites 60 references, 26 of which can be accessed free at http://www.jbc.org/content/288/37/26914.full.html#ref-list-1 Downloaded from http://www.jbc.org/ at Edinburgh University Library on September 13, 2013 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 288, NO. 37, pp. 26914–26925, September 13, 2013 © 2013 by The American Society for Biochemistry and Molecular Biology, Inc. Published in the U.S.A. Trypanosome Letm1 Protein Is Essential for Mitochondrial Potassium Homeostasis*□S Received for publication, June 19, 2013, and in revised form, July 23, 2013 Published, JBC Papers in Press, July 26, 2013, DOI 10.1074/jbc.M113.495119 Hassan Hashimi‡§1, Lindsay McDonald‡2, Eva Strˇíbrná‡, and Julius Lukesˇ‡§3 From the ‡Institute of Parasitology, Biology Centre, Czech Academy of Sciences and the §Faculty of Science, University of South Bohemia, 370 05 Cˇeské Budeˇjovice (Budweis), Czech Republic Background: Letm1 is a mitochondrial protein attributed disparate roles, including cation/proton antiport and translation. -

Loss of Frataxin Induces Iron Toxicity, Sphingolipid Synthesis, and Pdk1
1 Loss of Frataxin induces iron toxicity, sphingolipid synthesis, and 2 Pdk1/Mef2 activation, leading to neurodegeneration 3 4 Kuchuan Chen1, Guang Lin2, Nele A. Haelterman1, Tammy Szu-Yu Ho3, Tongchao Li1, Zhihong 5 Li2, Lita Duraine4, Brett H. Graham1, 2, Manish Jaiswal2, 4, Shinya Yamamoto1, 2, 5, Matthew N. 6 Rasband1, 3, and Hugo J. Bellen1, 2, 3, 4, 5, * 7 1Program in Developmental Biology, Baylor College of Medicine, Houston, TX 77030, USA 8 2Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA 9 3Department of Neuroscience, Baylor College of Medicine, Houston, TX 77030, USA 10 4Howard Hughes Medical Institute, Baylor College of Medicine, Houston, TX 77030, USA 11 5Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX 77030, 12 USA 13 14 Keywords: Friedreich ataxia; Frataxin; iron; mitochondria; sphingolipid; Pdk1; Mef2; lipid 15 droplets; photoreceptors; Drosophila 16 17 *Corresponding author: [email protected] 18 19 20 21 1 22 Abstract 23 Mutations in Frataxin (FXN) cause Friedreich’s ataxia (FRDA), a recessive 24 neurodegenerative disorder. Previous studies have proposed that loss of FXN causes 25 mitochondrial dysfunction, which triggers elevated reactive oxygen species (ROS) and leads to 26 the demise of neurons. Here we describe a ROS independent mechanism that contributes to 27 neurodegeneration in fly FXN mutants. We show that loss of frataxin homolog (fh) in Drosophila 28 leads to iron toxicity, which in turn induces sphingolipid synthesis and ectopically activates 3- 29 phosphoinositide dependent protein kinase-1 (Pdk1) and myocyte enhancer factor-2 (Mef2). 30 Dampening iron toxicity, inhibiting sphingolipid synthesis by Myriocin, or reducing Pdk1 or 31 Mef2 levels, all effectively suppress neurodegeneration in fh mutants. -

Functional Reconstitution of Mitochondrial Fe/S Cluster Synthesis on Isu1 Reveals the Involvement of Ferredoxin
ARTICLE Received 18 May 2014 | Accepted 19 Aug 2014 | Published 31 Oct 2014 DOI: 10.1038/ncomms6013 Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin Holger Webert1, Sven-Andreas Freibert1, Angelo Gallo2, Torsten Heidenreich1, Uwe Linne3, Stefan Amlacher4, Ed Hurt4, Ulrich Mu¨hlenhoff1, Lucia Banci2,5 & Roland Lill1,6,7 Maturation of iron–sulphur (Fe/S) proteins involves complex biosynthetic machinery. In vivo synthesis of [2Fe–2S] clusters on the mitochondrial scaffold protein Isu1 requires the cysteine desulphurase complex Nfs1-Isd11, frataxin, ferredoxin Yah1 and its reductase Arh1. The roles of Yah1–Arh1 have remained enigmatic, because they are not required for in vitro Fe/S cluster assembly. Here, we reconstitute [2Fe–2S] cluster synthesis on Isu1 in a reaction depending on Nfs1-Isd11, frataxin, Yah1, Arh1 and NADPH. Unlike in the bacterial system, frataxin is an essential part of Fe/S cluster biosynthesis and is required simultaneously and stoichiome- trically to Yah1. Reduced but not oxidized Yah1 tightly interacts with apo-Isu1 indicating a dynamic interaction between Yah1–apo-Isu1. Nuclear magnetic resonance structural studies identify the Yah1–apo-Isu1 interaction surface and suggest a pathway for electron flow from reduced ferredoxin to Isu1. Together, our study defines the molecular function of the ferre- doxin Yah1 and its human orthologue FDX2 in mitochondrial Fe/S cluster synthesis. 1 Institut fu¨r Zytobiologie und Zytopathologie, Philipps-Universita¨t Marburg, Robert-Koch-Strasse 6, 35032 Marburg, Germany. 2 CERM, Magnetic Resonance Center, University of Florence, Sesto Fiorentino, 50019 Florence, Italy. 3 Fachbereich Chemie, Philipps-Universita¨t Marburg, Hans-Meerwein-Street, 35043 Marburg, Germany. -

Diverse Entry Gates to Mitochondria Provide Insights Into the Evolution of Eukaryotes
1 Review Peeping at TOMs - Diverse entry gates to mitochondria provide insights into the evolution of eukaryotes Jan Mania, Chris Meisingerb and André Schneidera, 1 aDepartment of Chemistry and Biochemistry, University of Bern, Freiestrasse 3, CH- 3012 Bern, Switzerland bInstitut für Biochemie und Molekularbiologie, ZBMZ and BIOSS Centre for Biological Signalling Studies, Universität Freiburg, 79104 Freiburg, Germany 1 Correspondence to: [email protected] | downloaded: 13.3.2017 http://boris.unibe.ch/92438/ source: 2 Mitochondria are essential for eukaryotic life and more than 95% of their proteins are imported as precursors from the cytosol. The targeting signals for this post-translational import are conserved in all eukaryotes. However, this conservation does not hold true for the protein translocase of the mitochondrial outer membrane that serves as entry gate for essentially all precursor proteins. Only two of its subunits, Tom40 and Tom22, are conserved and thus likely were present in the last eukaryotic common ancestor. Tom7 is found in representatives of all supergroups except the Excavates. This suggests that it was added to the core of the translocase after the Excavates segregated from all other eukaryotes. A comparative analysis of the biochemically and functionally characterized outer membrane translocases of yeast, plants and trypanosomes, which represent three eukaryotic supergroups, shows that the receptors that recognize the conserved import signals differ strongly between the different systems. They present a remarkable example of convergent evolution at the molecular level. The structural diversity of the functionally conserved import receptors therefore provides insight into the early evolutionary history of mitochondria. Protein import distinguishes mitochondria from its endosymbiontic ancestor The origin of eukaryotic cells arguably is the most important transition in evolution besides the origin of life itself. -

030626 Mitochondrial Respiratory-Chain Diseases
The new england journal of medicine review article mechanisms of disease Mitochondrial Respiratory-Chain Diseases Salvatore DiMauro, M.D., and Eric A. Schon, Ph.D. From the Departments of Neurology (S.D., ore than a billion years ago, aerobic bacteria colonized E.A.S.) and Genetics and Development primordial eukaryotic cells that lacked the ability to use oxygen metabolical- (E.A.S.), Columbia University College of m Physicians and Surgeons, New York. Ad- ly. A symbiotic relationship developed and became permanent. The bacteria dress reprint requests to Dr. DiMauro at evolved into mitochondria, thus endowing the host cells with aerobic metabolism, a 4-420 College of Physicians and Surgeons, much more efficient way to produce energy than anaerobic glycolysis. Structurally, mito- 630 W. 168th St., New York, NY 10032, or at [email protected]. chondria have four compartments: the outer membrane, the inner membrane, the inter- membrane space, and the matrix (the region inside the inner membrane). They perform N Engl J Med 2003;348:2656-68. numerous tasks, such as pyruvate oxidation, the Krebs cycle, and metabolism of amino Copyright © 2003 Massachusetts Medical Society. acids, fatty acids, and steroids, but the most crucial is probably the generation of energy as adenosine triphosphate (ATP), by means of the electron-transport chain and the ox- idative-phosphorylation system (the “respiratory chain”) (Fig. 1). The respiratory chain, located in the inner mitochondrial membrane, consists of five multimeric protein complexes (Fig. 2B): reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase–ubiquinone oxidoreductase (complex I, approximately 46 sub- units), succinate dehydrogenase–ubiquinone oxidoreductase (complex II, 4 subunits), ubiquinone–cytochrome c oxidoreductase (complex III, 11 subunits), cytochrome c oxi- dase (complex IV, 13 subunits), and ATP synthase (complex V, approximately 16 sub- units). -

Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions
International Journal of Molecular Sciences Review Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions Margherita Protasoni 1 and Massimo Zeviani 1,2,* 1 Mitochondrial Biology Unit, The MRC and University of Cambridge, Cambridge CB2 0XY, UK; [email protected] 2 Department of Neurosciences, University of Padova, 35128 Padova, Italy * Correspondence: [email protected] Abstract: Mitochondria are ubiquitous intracellular organelles found in almost all eukaryotes and involved in various aspects of cellular life, with a primary role in energy production. The interest in this organelle has grown stronger with the discovery of their link to various pathologies, including cancer, aging and neurodegenerative diseases. Indeed, dysfunctional mitochondria cannot provide the required energy to tissues with a high-energy demand, such as heart, brain and muscles, leading to a large spectrum of clinical phenotypes. Mitochondrial defects are at the origin of a group of clinically heterogeneous pathologies, called mitochondrial diseases, with an incidence of 1 in 5000 live births. Primary mitochondrial diseases are associated with genetic mutations both in nuclear and mitochondrial DNA (mtDNA), affecting genes involved in every aspect of the organelle function. As a consequence, it is difficult to find a common cause for mitochondrial diseases and, subsequently, to offer a precise clinical definition of the pathology. Moreover, the complexity of this condition makes it challenging to identify possible therapies or drug targets. Keywords: ATP production; biogenesis of the respiratory chain; mitochondrial disease; mi-tochondrial electrochemical gradient; mitochondrial potential; mitochondrial proton pumping; mitochondrial respiratory chain; oxidative phosphorylation; respiratory complex; respiratory supercomplex Citation: Protasoni, M.; Zeviani, M.