The Molecular Evolution of B-Carbonic Anhydrase in Flaveria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Flaveria Pringlei (C3) Andflaveria Trinervia (C4) Under Nacl Stress
BIOLOGIAPLANTARUM 37 (1): 65-70, 1995 Flaveria pringlei (C3) and Flaveria trinervia (C4) under NaCI stress P. APEL, M. PEISKER, E. PFUNDEL and K.MISIHLE* Institut fi~r Pflanzengenetik und Kulturpflanzenforschung, Corrensstrafle 3, Gatersleben, D-06466, Germany Institut far Geophysik und Geologie der Universitdt Leipzig, Abt. Geochemm, Permoserstrafle 15, Letpztg, D-04303, Germany* Abstract The C 4 species Flaveria trinervia is obviously better adapted to saline environments than the C 3 species F pringlei, Treatment with 100 mM NaCl diminished crop growth rate in F. pringlei by 38 % but not in F. tnnervia. Under saline conditions, more assimilates were invested in leaf growth in F. trinervia but not in F. pringlei. Electrolyte concentration in F trinervia in control and salt treated plants is lower than in F. pringlei. Fluorescence data do not indicate a damage of PS 2 charge separation in both species. Whether the C 4 photosynthetic pathway in F. trinervia is responsible for the improved salt tolerance compared to F pringlei remains an open question. Key words: assimilates, clilorophyll, electrolytes, fluorescence, growth analysis, isotope discrimination, photosystem2, stomata Introduction The saline environments could have been an ecological niche which 'favoured genotypes exhibiting C 4 photosynthesis. Distribution of C 4 Atriplex species and their obvious preference of saline habitats support this assumption (Osmond et aL 1980). Powell (1978) reported in his monography of the genus Flaveria (Asteraceae) the salinity of natural habitats of F. australasica (C4), F. campestris (C4), F. trinervia (C4), F brownii (C4-1ike), F. chloraefolia (C3-C4), F. flondana (C3-C4) and F. oppositifolia (C3-C4). Although adaptation to salinity is a complex phenomenon (for review see e.g. -

Flaveria' Received for Publication February 21, 1986 R
Plant Physiol. (1986) 82, 211-217 0032-0889/86/82/0211/07/$0 1.00/0 Photosynthesis of F1 Hybrids between C4 and C3-C4 Species of Flaveria' Received for publication February 21, 1986 R. HAROLD BROWN, CAROLE L. BASSETT, RANDALL G. CAMERON, PHILIP T. EVANS, JOSEPH H. BOUTON*, CLANTON C. BLACK, JR., LEONEL O'REILLY STERNBERG, AND MICHAEL J. DENIRO Department ofAgronomy (R.H.B., P.T.E., J.H.B., R.G.C.) and Department ofBiochemistry (C.C.B.), University ofGeorgia, Athens, Georgia 30602; Richard Russell Research Center, United States Department ofAgriculture, Agricultural Research Service, Athens, Georgia (C.L.B.); Department of Biology, University ofMiami, Coral Gables, Florida 33124 (L.O'R.S.); and Department ofEarth and Space Sciences and Archaeology Program, University ofCalifornia, Los Angeles, California 90024 (M.J.D.) ABSTRACT not been found which are closely enough related to produce highly fertile offspring. But species have been discovered in Photosynthetic characteristics were studied in several F, hybrids be- several genera which have photosynthetic and leaf anatomical tween C4 and C3-C4 species of Flaveria. Stable carbon isotope ratios, 02 characteristics intermediate between C3 and C4 species (C3-C4) inhibition of apparent photosynthesis, and phosphoenolpyruvate carbox- (11, 20, 22). These intermediate characteristics may indicate ylase activities in the hybrids were similar to the means for the parents. closer phylogenetic relationships with C4 species and greater Values of CO2 compensation concentrations were nearer to those of the success in hybridization. Because C3-C4 species in most cases fix C4 parent and apparent photosynthesis was below that of both parents, CO2 exclusively by the C3 cycle (10, 11, 20, 29), hybrids between being only 60 and 74% of that of the lowest (C3-C4) parent in two C4 and C3-C4 species may be useful in understanding genetic experiments. -

Anti-CA1 / Carbonic Anhydrase 1 Antibody (ARG65670)
Product datasheet [email protected] ARG65670 Package: 100 μl, 50 μl anti-CA1 / Carbonic Anhydrase 1 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes CA1 / Carbonic Anhydrase 1 Tested Reactivity Ms, Rat Tested Application WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name CA1 / Carbonic Anhydrase 1 Antigen Species Human Immunogen Full length fusion protein of Human CA1. Conjugation Un-conjugated Alternate Names Carbonic anhydrase I; EC 4.2.1.1; Carbonate dehydratase I; Carbonic anhydrase B; Car1; HEL-S-11; CA-I; Carbonic anhydrase 1; CAB Application Instructions Application table Application Dilution WB 1:500 - 1:2000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control WB: Mouse liver tissue Calculated Mw 29 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer PBS (pH 7.3), 0.05% Sodium azide and 50% Glycerol Preservative 0.05% Sodium azide Stabilizer 50% Glycerol Concentration 0.9 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. www.arigobio.com 1/2 Bioinformation Gene Symbol CA1 Gene Full Name carbonic anhydrase I Background Carbonic anhydrases (CAs) are a large family of zinc metalloenzymes that catalyze the reversible hydration of carbon dioxide. -

Flórula Vascular De La Sierra De Catorce Y Territorios Adyacentes, San Luis Potosi, México
Acta Botanica Mexicana 78: 1-38 (2007) FLÓRULA VASCULAR DE LA SIERRA DE CATORCE Y TERRITORIOS ADYACENTES, SAN LUIS POTOSI, MÉXICO ONÉSIMO GONZÁLEZ COSTILLA1,2, JOAQUÍN GIMÉNEZ DE AZCÁRATE3, JOSÉ GARCÍA PÉREZ1 Y JUAN RogELIO AGUIRRE RIVERA1 1Universidad Autónoma de San Luis Potosí, Instituto de Investigación de Zonas Desérticas, Altair 200, Fraccionamiento El Llano, Apdo. postal 504, 78377 San Luis Potosí, México. 2Universidad Complutense de Madrid, Departamento de Biología Vegetal II, Facultad de Farmacia, Madrid, España. [email protected] 3Universidad de Santiago de Compostela, Departamento de Botánica, Escuela Politécnica Superior, 27002 Lugo, España. RESUMEN La Sierra de Catorce, localizada en el norte del estado de San Luis Potosí, reúne algunas de las principales cimas del Desierto Chihuahuense cuyas cotas superan los 3000 metros. Ello ha favorecido que la Sierra sea una importante área de diversificación de la flora y las fitocenosis de dicha ecorregión. A partir del estudio fitosociológico de la vegetación del territorio, que se está realizando desde 1999, se ha obtenido un catálogo preliminar de su flora. Hasta el momento la lista de plantas vasculares está conformada por 526 especies y cuatro taxa infraespecíficos, agrupados en 293 géneros y 88 familias. Las familias y géneros mejor representados son Asteraceae, Poaceae, Cactaceae, Fabaceae, Fagaceae y Lamiaceae, así como Quercus, Opuntia, Muhlenbergia, Salvia, Agave, Bouteloua y Dyssodia, respectivamente. Asimismo se señalan los tipos de vegetación representativos del área que albergan los diferentes taxa. Por último, con base en diferentes listas de flora amenazada, se identificaron las especies incluidas en alguna de las categorías reconocidas. Palabras clave: Desierto Chihuahuense, estudio fitosociológico, flora, flora ame- nazada, México, San Luis Potosí, Sierra de Catorce. -

Neglected Functions of TFCP2/TFCP2L1/UBP1 Transcription Factors May Offer Valuable Insights Into Their Mechanisms of Action
International Journal of Molecular Sciences Review Neglected Functions of TFCP2/TFCP2L1/UBP1 Transcription Factors May Offer Valuable Insights into Their Mechanisms of Action Agnieszka Taracha, Grzegorz Kotarba and Tomasz Wilanowski * Laboratory of Signal Transduction, Nencki Institute of Experimental Biology of Polish Academy of Sciences, 3 Pasteur St., 02-093 Warsaw, Poland; [email protected] (A.T.); [email protected] (G.K.) * Correspondence: [email protected]; Tel.: +48-22-5892-311 Received: 21 August 2018; Accepted: 19 September 2018; Published: 20 September 2018 Abstract: In recent years, the TFCP2 (transcription factor cellular promoter 2)/TFCP2L1 (TFCP2- like 1)/UBP1 (upstream binding protein 1) subfamily of transcription factors has been attracting increasing attention in the scientific community. These factors are very important in cancer, Alzheimer’s disease, and other human conditions, and they can be attractive targets for drug development. However, the interpretation of experimental results is complicated, as in principle, any of these factors could substitute for the lack of another. Thus, studying their hitherto little known functions should enhance our understanding of mechanisms of their functioning, and analogous mechanisms might govern their functioning in medically relevant contexts. For example, there are numerous parallels between placental development and cancer growth; therefore, investigating the roles of TFCP2, TFCP2L1, and UBP1 in the placenta may help us better understand their functioning in cancer, as is evidenced by the studies of various other proteins and pathways. Our review article aims to call the attention of the scientific community to these neglected functions, and encourage further research in this field. -

Flaveria Bidentis (L.) Kuntze (Asteraceae), a Newly Naturalized Plant in Taiwan
林業研究季刊 30(4) : 23-詣 , 2008 23 蚯盔盤益 臺灣新歸化菊科植物-黃頂菊 曾彥學 1 劉靜愉 2 嚴新富 3 彭鏡毅 4 , 5 [摘要】本文首次報導原產北美洲南部,目前已歸化於台灣嘉義縣鱉鼓沿海附近的黃頂菊(菊科) , 描述其形態特徵、地理分布及生育地環境,並提供彩色圖片與線繪圖以資辨識 。 作者等於 1987 年 即已發現黃頂菊族群, 2008 年再進行調查時發現其野外族群穩定成長 。 黃頂菊為臺灣新歸化植物, 本屬亦為臺灣新記錄屬 。 [關鍵詞]菊科、黃頂菊、歸化植物、臺灣 Research paper Flaveria bidentis (L.) Kuntze (Asteraceae), a Newly Naturalized Plant in Taiwan Yen-Hsueh Tseng1 Chíng-Yu Líou2 Hsin-Fu Yen3 Ching-I Peng4,5 <Abstract) We document the naturalization of Flaveria bidentis (L.) Kuntze (Asteraceae) in southem Taiwan. This is one ofthe many cases ofNew World plants invading Taiwan. A taxonomic treatment, line drawings, and color photographs of this species 企om the wild are provided to aid in identification of this alien plant. The colony of F bidentis was first observed in Taiwan in1987. During our field survey in 2008 we witnessed that the wild populations has adapted to the coast of Chiayi County. (Key words] Asteraceae, Flaveria bident肉, Taxonomy, Naturalized plant, Taiwan 1.團立中興大學森林學系, 402 台中市南區團光路 250 號,台灣 Department ofForestry, National Chung-Hsing University, Taichung 402, Taiwan. 2 . 特有生物研究保育 中心棲地生態組, 552 南投縣集集鎮民生東路 1 號,台灣 Department ofHabitat and Ecology, Taiwan Endemic Species Research Institute , Nantou 552, Taiwan. 3. 國立自然科學博物館植物學組, 404 台中市館前路 1 號,臺 j彎 Department ofBotany, National Museum ofNatural Science, 1, Guancian Rd. , Taichung 404, Taiwan. 4 . 中央研究院生物多樣性研究中心, 115 台北市研究院路 2 段 128 號,台灣 Herbarium (HAST), Research Center for Biodiversity, Academia Sinica, Nankang, Taipei 115, Taiwan. 5 . 通訊作者: Email:[email protected] Corresponding author. EmaiJ : [email protected] 24 臺灣新歸化菊科植物-黃頂菊 INTRODUCTION phyllaries, with 2 additional small exterior ones. In recent years, many species of Asteraceae Receptacle small, glabrous. -
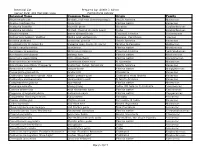
Botanical Name Common Name Origin Family
Botanical List Prepared by: Arielle J. Simon Corner Park: 401 Hampton Lane Horticultural Advisor Botanical Name Common Name Origin Family Abutilon pictum fireball, red vein flowering maple South America Malvaceae Acacia choriophylla cinnecord Florida native Fabaceae Acalypha hispida chenille plant Oceania Euphorbiaceae Acalypha pendula firetail, trailing chenille plant Cuba Euphorbiaceae Aloysia virgata sweet almond bush Tropical America Verbenaceae Anthurium hookeri 'Ruffles' bird's nest anthurium Guyana, Caribbean Araceae Arachis glabrata perennial peanut South America Fabaceae Arachnothryx leucophylla Panama rose, huele de noche Mexico to Panama Rubiaceae Ardisia escallonioides marlberry Florida native Myrsinaceae Asclepias curassavica Mexican milkweed Tropical America Asclepiadaceae Blechnum serrulatum swamp fern Florida native Blechnaceae Bourreria cassinifolia little strongback Florida native Boraginaceae Brachychiton acerifolius Australian flame tree N Australia Malvaceae Brunfelsia pauciflora 'Compacta' yesterday, today, tomorrow South America Fabaceae Byrsonima lucida locust-berry Florida native Malpighiaceae Caesalpinia granadillo bridal veil Venezuela Fabaceae Calliandra haematocephala 'Alba' white powder-puff Cultivated from Bolivia Fabaceae Calliandra surinamensis pink powder-puff N South America Fabaceae Calyptranthes pallens spicewood Florida native Myrtaceae Cananga odorata ylang-ylang India, SE Asia to N Australia Annonaceae Canella winterana wild cinnamon bark Florida native Canellaceae Capparis cynophallophora Jamaican -

The Observed Alteration in BCL2 Expression Following Lithium
www.nature.com/scientificreports OPEN The observed alteration in BCL2 expression following lithium treatment is infuenced by the Received: 26 August 2016 Accepted: 26 March 2018 choice of normalization method Published: xx xx xxxx Damri Odeya1,2, Agam Galila1,2 & Toker Lilah3,4 Upregulation of B-cell CLL/lymphoma (BCL)2 expression following lithium treatment is seemingly well established and has been related to the neuroprotective property of the drug. However, while demonstrated by some (but not all) studies based on low-throughput techniques (e.g. qPCR) this efect is not refected in high-throughput studies, such as microarrays and RNAseq. This manuscript presents a systematic review of currently available reports of lithium’s efect on BCL2 expression. To our surprise, we found that the majority of the literature does not support the efect of lithium onBCL 2 transcript or protein levels. Moreover, among the positive reports, several used therapeutically irrelevant lithium doses while others lack statistical power. We also noticed that numerous low-throughput studies normalized the signal using genes/proteins afected by lithium, imposing possible bias. Using wet bench experiments and reanalysis of publicly available microarray data, here we show that the reference gene chosen for normalization critically impacts the outcome of qPCR analyses of lithium’s efect on BCL2 expression. Our fndings suggest that experimental results might be severely afected by the choice of normalizing genes, and emphasize the need to re- evaluate stability of these genes in the context of the specifc experimental conditions. Upregulation of B-cell CLL/lymphoma (BCL)2 transcript following chronic lithium treatment is considered to be well established1. -

Plant List: M.E. Depalma Park Picture Botanical Name Common Name Picture Botanical Name Common Name Salvia Coccinea Tropical
Plant List: M.E. DePalma Park Picture Botanical Name Picture Botanical Name Common Name Common Name Salvia coccinea Muhlenbergia capillaris Tropical Sage (red) pink Florida Muhly Grass Blooms: Year-round Blooms: Mid Summer - Fall Attracts Butterflies, Hummingbirds and Painted Buntings Florida Native Florida Native Psychotria nervosa Arachis glabrata Wild Coffee Perennial peanut Blooms: Spring-Summer Fruits: Fall Blooms: Year-round Berries are a food source for birds Native to Argentina, Brazil and Paraguay Male Butterflies need the alkaloids found in this plant, in order to mature sexually. Florida Native Callicarpa americana Gaillardia pulchella Beautyberry Blanket Flower Blooms: late Summer Fruits: Fall Blooms: Year-round Berries are a food source for birds Butterfly nectar plant Florida Native Florida Native Chrysobalanus icaco Stachytarpheta jamaicensis Cocoplum Blue Porterweed Blooms: Spring-Summer Fruits: Fall Blooms: Year-round Berries are a food source for birds Butterfly nectar plant Florida Native Florida Native Asclepias curassavica Pentas lanceolata Mexican Butterfly Weed, Blood-flower or Scarlet Milkweed Egyptian starflower Blooms: Year-round Blooms: Year-round Butterfly nectar plant for Butterflies especially Queens and These may be red, white, lavender, purple, or shades Monarchs of pink. Some are two-toned. All are extremely Monarch larval plant attractive to butterflies, and the red and dark pink varieties delight hummingbirds. Butterfly nectar plant Mimosa strigillosa Loropetalum Sensitive Plant Chinese Fringe Flower Blooms: Year-round Butterfly nectar plant Butterfly nectar plant Native to Japan and southeastern Asia including Florida Native groundcover southern China. Coreopsis_leavenworthii Coreopsis lanceolata Tickseed Lance leafed coreopsis Blooms: Year-round Butterfly nectar plant Butterfly nectar plant Florida Native Florida Native Lantana involucrata. -

Engineered Type 1 Regulatory T Cells Designed for Clinical Use Kill Primary
ARTICLE Acute Myeloid Leukemia Engineered type 1 regulatory T cells designed Ferrata Storti Foundation for clinical use kill primary pediatric acute myeloid leukemia cells Brandon Cieniewicz,1* Molly Javier Uyeda,1,2* Ping (Pauline) Chen,1 Ece Canan Sayitoglu,1 Jeffrey Mao-Hwa Liu,1 Grazia Andolfi,3 Katharine Greenthal,1 Alice Bertaina,1,4 Silvia Gregori,3 Rosa Bacchetta,1,4 Norman James Lacayo,1 Alma-Martina Cepika1,4# and Maria Grazia Roncarolo1,2,4# Haematologica 2021 Volume 106(10):2588-2597 1Department of Pediatrics, Division of Stem Cell Transplantation and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 2Stanford Institute for Stem Cell Biology and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 3San Raffaele Telethon Institute for Gene Therapy, Milan, Italy and 4Center for Definitive and Curative Medicine, Stanford School of Medicine, Stanford, CA, USA *BC and MJU contributed equally as co-first authors #AMC and MGR contributed equally as co-senior authors ABSTRACT ype 1 regulatory (Tr1) T cells induced by enforced expression of interleukin-10 (LV-10) are being developed as a novel treatment for Tchemotherapy-resistant myeloid leukemias. In vivo, LV-10 cells do not cause graft-versus-host disease while mediating graft-versus-leukemia effect against adult acute myeloid leukemia (AML). Since pediatric AML (pAML) and adult AML are different on a genetic and epigenetic level, we investigate herein whether LV-10 cells also efficiently kill pAML cells. We show that the majority of primary pAML are killed by LV-10 cells, with different levels of sensitivity to killing. Transcriptionally, pAML sensitive to LV-10 killing expressed a myeloid maturation signature. -

Asteraceae: Astereae), an Endemic Shrub of the Galapagos Islands Nicole Genet Andrus Florida International University
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 7-24-2002 The origin, phylogenetics and natural history of darwiniothamnus (Asteraceae: Astereae), an endemic shrub of the Galapagos Islands Nicole Genet Andrus Florida International University DOI: 10.25148/etd.FI14032319 Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Biology Commons Recommended Citation Andrus, Nicole Genet, "The origin, phylogenetics and natural history of darwiniothamnus (Asteraceae: Astereae), an endemic shrub of the Galapagos Islands" (2002). FIU Electronic Theses and Dissertations. 1290. https://digitalcommons.fiu.edu/etd/1290 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida THE ORIGIN, PHYLOGENETICS AND NATURAL HISTORY OF DARWINIOTHAMNUS (ASTERACEAE: ASTEREAE), AN ENDEMIC SHRUB OF THE GALAPAGOS ISLANDS A thesis submitted in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE in BIOLOGY by Nicole Genet Andrus 2002 To: Dean Arthur W. Herriott College of Arts and Sciences This thesis, written by Nicole Genet Andrus, and entitled The Origin, Phylogenetics and Natural History of Darwiniothamnus (Asteraceae: Astereae), an Endemic Shrub of the Galapagos Islands, having been approved in respect to style and intellectual content, is referred to you for judgment. We have read this thesis and recommend that it be approved. Alan Tye Susan Koptur Carl Lewis Javiefr acisco-Ortega, Major Professor Date of Defense: July 24, 2002 The thesis of Nicole Genet Andrus is approved. -

Expression of Carbonic Anhydrase II
Biochemical Genetics, VoL 33, Nos. 11/12, 1995 Expression of Carbonic Anhydrase II (CA II) Promoter-Reporter Fusion Genes in Multiple Tissues of Transgenic Mice Does Not Replicate Normal Patterns of Expression Indicating Complexity of CA II Regulation in Vivo Robert P. Erickson, 1,2,4 Judy Grimes, 1 Patrick J. Venta, 3 and Richard E. Tashian 3 Received 6 June 1995 Final5 Sept. 1995 Although the proximal, 5' 115 bp of the human carbonic anhydrase II (CA II) gene was sufficient for expression of a reporter gene in some transfected cell lines, we found previously that 1100 bp of this promoter (or 500 bp of the mouse CA II promoter) was not sufficient for expression in transgenic mice. We have now studied the expression of linked reporter genes in mice transgenic for either (1) l 1 kb of the human 5' promoter or (2) 8 kb of the human 5' promoter with mouse sequences from the first exon, part of the first intron (since a CpG island spans this region), and the 3' sequences of the gene. Expression was found in both cases, but the tissue specificity was not appropriate for CA II. Although there was a difference in the sensitivity of the assays used, the frst construct led to expression in many tissues, while the second construct was expressed only in spleen. These findings indicate considerable complexity of DNA control regions for in vivo CA II expression. KEY WORDS: transgenic mice; carbonic anhydrase; promoter analysis; transcription; DNA control regions. Angel Charity for Children-Wings for Genetic Research, Steele Memorial Children's Re- search Center, Department of Pediatrics, University of Arizona, Tucson, Arizona.