Investigating Wood Decaying Fungi Diversity in Central Siberia, Russia Using ITS Sequence Analysis and Interaction with Host Trees
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Survey of Fungi at the University of Wisconsin-Waukesha Field Station
University of Wisconsin Milwaukee UWM Digital Commons Field Station Bulletins UWM Field Station Spring 1993 A survey of fungi at the University of Wisconsin- Waukesha Field Station Alan D. Parker University of Wisconsin-Waukesha Follow this and additional works at: https://dc.uwm.edu/fieldstation_bulletins Part of the Forest Biology Commons, and the Zoology Commons Recommended Citation Parker, A.D. 1993 A survey of fungi at the University of Wisconsin-Waukesha Field Station. Field Station Bulletin 26(1): 1-10. This Article is brought to you for free and open access by UWM Digital Commons. It has been accepted for inclusion in Field Station Bulletins by an authorized administrator of UWM Digital Commons. For more information, please contact [email protected]. A Survey of Fungi at the University of Wisconsin-Waukesha Field Station Alan D. Parker Department of Biological Sciences University of Wisconsin-Waukesha Waukesha, Wisconsin 53188 Introduction The University of Wisconsin-Waukesha Field Station was founded in 1967 through the generous gift of a 98 acre farm by Ms. Gertrude Sherman. The facility is located approximately nine miles west of Waukesha on Highway 18, just south of the Waterville Road intersection. The site consists of rolling glacial deposits covered with old field vegetation, 20 acres of xeric oak woods, a small lake with marshlands and bog, and a cold water stream. Other communities are being estab- lished as a result of restoration work; among these are mesic prairie, oak opening, and stands of various conifers. A long-term study of higher fungi and Myxomycetes, primarily from the xeric oak woods, was started in 1978. -
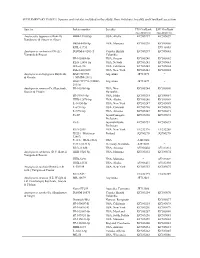
Suppl Table I
SUPPLEMENTARY TABLE I. Species and isolates included in the study, their vouchers, locality and GenBank accession Species Isolate number Locality ITS GenBank LSU GenBank accession no. accession no. Amylocystis lapponica (Romell) HHB-13400-Sp USA, Alaska KC585237 KC585059 Bondartsev & Singer ex Singer OKM-4418-Sp USA, Montana KC585238 KC585060 KHL-11755 − EU118603 Amyloporia carbonica (Overh.) DAOM-F-8281-T Canada, British KC585239 KC585061 Vampola & Pouzar Columbia FP-105585-Sp USA, Oregon KC585240 KC585062 RLG-12496-Sp USA, Nevada KC585241 KC585063 Wilcox-96 USA, California KC585242 KC585064 Zabel-40-GLN USA, New York KC585243 KC585065 Amyloporia nothofaginea Rajchenb. BAFC519794 Argentina JF713078 − & Gorjón (=MMBP-2011) BAFC519796 (=MBP- Argentina JF713079 − 2011a) Amyloporia sinuosa (Fr.) Rajchenb., FP-105386-Sp USA, New KC585244 KC585066 Gorjón & Pildain Hampshire FP-94464-Sp USA, Idaho KC585245 KC585067 HHB-12878-Sp USA, Alaska KC585246 KC585068 L-14130-Sp USA, New York KC585247 KC585069 L-6192-Sp USA, Colorado KC585248 KC585070 L-9792-Sp USA, Arizona KC585249 KC585071 Pa-3C JapanYamagata KC585250 KC585072 Prefecture Pa-3e JapanShizuoka KC585251 KC585073 Prefecture RLG-2538 USA, New York EU232196 EU232288 X725 (=Miettinen- Finland JQ700270 JQ700270 12407) P-115 (=RLG-2538) USA AJ416068 − P-211 (G-214) Germany, Karlsrube AJ345011 − RLG-1182R USA, Arizona AY966450 AY333831 Amyloporia sitchensis (D.V. Baxter) HHB-5320-Sp USA, Montana KC585252 KC585074 Vampola & Pouzar HHB-5298 USA, Montana − AY333829 HHB-12513 USA, Alaska AY966451 AY333830 -

Major Clades of Agaricales: a Multilocus Phylogenetic Overview
Mycologia, 98(6), 2006, pp. 982–995. # 2006 by The Mycological Society of America, Lawrence, KS 66044-8897 Major clades of Agaricales: a multilocus phylogenetic overview P. Brandon Matheny1 Duur K. Aanen Judd M. Curtis Laboratory of Genetics, Arboretumlaan 4, 6703 BD, Biology Department, Clark University, 950 Main Street, Wageningen, The Netherlands Worcester, Massachusetts, 01610 Matthew DeNitis Vale´rie Hofstetter 127 Harrington Way, Worcester, Massachusetts 01604 Department of Biology, Box 90338, Duke University, Durham, North Carolina 27708 Graciela M. Daniele Instituto Multidisciplinario de Biologı´a Vegetal, M. Catherine Aime CONICET-Universidad Nacional de Co´rdoba, Casilla USDA-ARS, Systematic Botany and Mycology de Correo 495, 5000 Co´rdoba, Argentina Laboratory, Room 304, Building 011A, 10300 Baltimore Avenue, Beltsville, Maryland 20705-2350 Dennis E. Desjardin Department of Biology, San Francisco State University, Jean-Marc Moncalvo San Francisco, California 94132 Centre for Biodiversity and Conservation Biology, Royal Ontario Museum and Department of Botany, University Bradley R. Kropp of Toronto, Toronto, Ontario, M5S 2C6 Canada Department of Biology, Utah State University, Logan, Utah 84322 Zai-Wei Ge Zhu-Liang Yang Lorelei L. Norvell Kunming Institute of Botany, Chinese Academy of Pacific Northwest Mycology Service, 6720 NW Skyline Sciences, Kunming 650204, P.R. China Boulevard, Portland, Oregon 97229-1309 Jason C. Slot Andrew Parker Biology Department, Clark University, 950 Main Street, 127 Raven Way, Metaline Falls, Washington 99153- Worcester, Massachusetts, 01609 9720 Joseph F. Ammirati Else C. Vellinga University of Washington, Biology Department, Box Department of Plant and Microbial Biology, 111 355325, Seattle, Washington 98195 Koshland Hall, University of California, Berkeley, California 94720-3102 Timothy J. -

The Use of Algae and Fungi for Removal of Pharmaceuticals by Bioremediation and Biosorption Processes: a Review
Review The Use of Algae and Fungi for Removal of Pharmaceuticals by Bioremediation and Biosorption Processes: A Review Andreia Silva, Cristina Delerue-Matos, Sónia A. Figueiredo and Olga M. Freitas * REQUIMTE/LAQV, Instituto Superior de Engenharia do Porto, Rua Dr. António Bernardino de Almeida 431, 4200-072 Porto, Portugal * Correspondence: [email protected] Received: 3 July 2019; Accepted: 25 July 2019; Published: 27 July 2019 Abstract: The occurrence and fate of pharmaceuticals in the aquatic environment is recognized as one of the emerging issues in environmental chemistry. Conventional wastewater treatment plants (WWTPs) are not designed to remove pharmaceuticals (and their metabolites) from domestic wastewaters. The treatability of pharmaceutical compounds in WWTPs varies considerably depending on the type of compound since their biodegradability can differ significantly. As a consequence, they may reach the aquatic environment, directly or by leaching of the sludge produced by these facilities. Currently, the technologies under research for the removal of pharmaceuticals, namely membrane technologies and advanced oxidation processes, have high operation costs related to energy and chemical consumption. When chemical reactions are involved, other aspects to consider include the formation of harmful reaction by-products and the management of the toxic sludge produced. Research is needed in order to develop economic and sustainable treatment processes, such as bioremediation and biosorption. The use of low-cost materials, such as biological matrices (e.g., algae and fungi), has advantages such as low capital investment, easy operation, low operation costs, and the non-formation of degradation by-products. An extensive review of existing research on this subject is presented. -

Field Guide to Common Macrofungi in Eastern Forests and Their Ecosystem Functions
United States Department of Field Guide to Agriculture Common Macrofungi Forest Service in Eastern Forests Northern Research Station and Their Ecosystem General Technical Report NRS-79 Functions Michael E. Ostry Neil A. Anderson Joseph G. O’Brien Cover Photos Front: Morel, Morchella esculenta. Photo by Neil A. Anderson, University of Minnesota. Back: Bear’s Head Tooth, Hericium coralloides. Photo by Michael E. Ostry, U.S. Forest Service. The Authors MICHAEL E. OSTRY, research plant pathologist, U.S. Forest Service, Northern Research Station, St. Paul, MN NEIL A. ANDERSON, professor emeritus, University of Minnesota, Department of Plant Pathology, St. Paul, MN JOSEPH G. O’BRIEN, plant pathologist, U.S. Forest Service, Forest Health Protection, St. Paul, MN Manuscript received for publication 23 April 2010 Published by: For additional copies: U.S. FOREST SERVICE U.S. Forest Service 11 CAMPUS BLVD SUITE 200 Publications Distribution NEWTOWN SQUARE PA 19073 359 Main Road Delaware, OH 43015-8640 April 2011 Fax: (740)368-0152 Visit our homepage at: http://www.nrs.fs.fed.us/ CONTENTS Introduction: About this Guide 1 Mushroom Basics 2 Aspen-Birch Ecosystem Mycorrhizal On the ground associated with tree roots Fly Agaric Amanita muscaria 8 Destroying Angel Amanita virosa, A. verna, A. bisporigera 9 The Omnipresent Laccaria Laccaria bicolor 10 Aspen Bolete Leccinum aurantiacum, L. insigne 11 Birch Bolete Leccinum scabrum 12 Saprophytic Litter and Wood Decay On wood Oyster Mushroom Pleurotus populinus (P. ostreatus) 13 Artist’s Conk Ganoderma applanatum -

Oxalic Acid Degradation by a Novel Fungal Oxalate Oxidase from Abortiporus Biennis Marcin Grąz1*, Kamila Rachwał2, Radosław Zan2 and Anna Jarosz-Wilkołazka1
Vol. 63, No 3/2016 595–600 http://dx.doi.org/10.18388/abp.2016_1282 Regular paper Oxalic acid degradation by a novel fungal oxalate oxidase from Abortiporus biennis Marcin Grąz1*, Kamila Rachwał2, Radosław Zan2 and Anna Jarosz-Wilkołazka1 1Department of Biochemistry, Maria Curie-Skłodowska University, Lublin, Poland; 2Department of Genetics and Microbiology, Maria Curie-Skłodowska University, Lublin, Poland Oxalate oxidase was identified in mycelial extracts of a to formic acid and carbon dioxide (Mäkelä et al., 2002). basidiomycete Abortiporus biennis strain. Intracellular The degradation of oxalate via action of oxalate oxidase enzyme activity was detected only after prior lowering (EC 1.2.3.4), described in our study, is atypical for fun- of the pH value of the fungal cultures by using oxalic or gi and was found predominantly in higher plants. The hydrochloric acids. This enzyme was purified using size best characterised oxalate oxidase originates from cereal exclusion chromatography (Sephadex G-25) and ion-ex- plants (Dunwell, 2000). Currently, only three oxalate oxi- change chromatography (DEAE-Sepharose). This enzyme dases of basidiomycete fungi have been described - an exhibited optimum activity at pH 2 when incubated at enzyme from Tilletia contraversa (Vaisey et al., 1961), the 40°C, and the optimum temperature was established at best characterised so far enzyme from Ceriporiopsis subver- 60°C. Among the tested organic acids, this enzyme ex- mispora (Aguilar et al., 1999), and an enzyme produced by hibited specificity only towards oxalic acid. Molecular Abortiporus biennis (Grąz et al., 2009). The enzyme from mass was calculated as 58 kDa. The values of Km for oxa- C. -

Hori Et Al 2013.Pdf
Mycologia, 105(6), 2013, pp. 1412–1427. DOI: 10.3852/13-072 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay Chiaki Hori cellulases belonging to families GH6, GH7, GH9 Department of Biomaterials Sciences, Graduate School of and carbohydrate-binding module family CBM1 are Agricultural and Life Sciences, University of Tokyo, l-l-l, lacking in genomes of brown-rot polyporales. In Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan, and Institute for Microbial and Biochemical Technology, addition, the presence of CDH and the expansion Forest Products Laboratory, 1 Gifford Pinchot Drive, of LPMO were observed only in white-rot genomes. Madison, Wisconsin 53726 Indeed, GH6, GH7, CDH and LPMO peptides were identified only in white-rot polypores. Genes encod- Jill Gaskell ing aldose 1-epimerase (ALE), previously detected Institute for Microbial and Biochemical Technology, Forest Products Laboratory, 1 Gifford Pinchot Drive, with CDH and cellulases in the culture filtrates, also Madison, Wisconsin 53726 were identified in white-rot genomes, suggesting a physiological connection between ALE, CDH, cellu- Kiyohiko Igarashi lase and possibly LPMO. For hemicellulose degrada- Masahiro Samejima tion, genes and peptides corresponding to GH74 Department of Biomaterials Sciences, Graduate School of Agricultural and Life Sciences, University of Tokyo, l-l-l, xyloglucanase, GH10 endo-xylanase, GH79 b-glucu- Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan ronidase, CE1 acetyl xylan esterase and CE15 glucur- onoyl methylesterase were significantly increased in David Hibbett white-rot genomes compared to brown-rot genomes. -

Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia Radiculosa
Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia radiculosa Juliet D. Tang,a Andy D. Perkins,b Tad S. Sonstegard,c Steven G. Schroeder,c Shane C. Burgess,d and Susan V. Diehla Forest Products, Mississippi State University, Mississippi State, Mississippi, USAa; Computer Science and Engineering, Mississippi State University, Mississippi State, Mississippi, USAb; USDA ARS Bovine Functional Genomics Laboratory, Beltsville, Maryland, USAc; and Institute for Genomics, Biocomputing, and Biotechnology, Mississippi State University, Mississippi State, Mississippi, USAd The feasibility of short-read sequencing for genomic analysis was demonstrated for Fibroporia radiculosa, a copper-tolerant fungus that causes brown rot decay of wood. The effect of read quality on genomic assembly was assessed by filtering Illumina GAIIx reads from a single run of a paired-end library (75-nucleotide read length and 300-bp fragment size) at three different stringency levels and then assembling each data set with Velvet. A simple approach was devised to determine which filter strin- gency was “best.” Venn diagrams identified the regions containing reads that were used in an assembly but were of a low-enough quality to be removed by a filter. By plotting base quality histograms of reads in this region, we judged whether a filter was too stringent or not stringent enough. Our best assembly had a genome size of 33.6 Mb, an N50 of 65.8 kb for a k-mer of 51, and a maximum contig length of 347 kb. Using GeneMark, 9,262 genes were predicted. TargetP and SignalP analyses showed that among the 1,213 genes with secreted products, 986 had motifs for signal peptides and 227 had motifs for signal anchors. -

Climacodon Pulcherrimus a Badly Known Tropical Species, Present in Europe
Cryptogamie,Mycologie, 2007, 28 (1): 3-11 © 2007 Adac. Tous droits réservés Climacodon pulcherrimus a badly known tropical species, present in Europe Gabriel MORENOa*, María Natividad BLANCOa , Ibai OLARIAGAb &Julia CHECAa a Dpto. Biología Vegetal, Fac. Biología. Univ. de Alcalá, E-28871,Alcalá de Henares, Madrid. e-mail: [email protected] b Dpto. Biología Vegetal y Ecología (Botánica). Fac. Ciencias y Tecnología. Campus de Leioa. Univ. del País Vasco. Apartado 644. E-48080 Bilbao. Abstract – Climacodon pulcherrimus,apolymorphous species with a large distribution and habitat, is described macro- and microscopically. Its uncertain taxonomic position has led to the description of many synonymous species and placement in different genera. The type species of Hydnum pulcherrimum Berk. &M.A. Curtis is examined for the first time and it is compared with other collections from Malay Peninsula, Pakistan, USA and Spain. The study of Spanish collections enlarges the distribution to the South of Europe. Basidiomycota / Meruliaceae / Climacodon / Donkia / Hydnum / systematics / chorology / taxonomy INTRODUCTION In the last four years, we have collected some basidiomata of a saprophyte fungus with a basidioma of medium size, normally dimidiate with trametoid appearance and hydnoid hymenophore. Microscopically it is characterized by the presence of double or multiple clamp connections, ellipsoid basidiospores and absence of cystidia. Finally, we could determine it, with some difficulties, as a member of the genus Climacodon P. Karst., belonging to the family Meruliaceae P. Karst., order Polyporales Gäum. (Kirk et al., 2001). This genus includes species with conspicuous cystidia and hyphae with single clamp connections, with the only exception of C. pulcherrimus (Berk. -

A New Species of Antrodia (Basidiomycota, Polypores) from China
Mycosphere 8(7): 878–885 (2017) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/8/7/4 Copyright © Guizhou Academy of Agricultural Sciences A new species of Antrodia (Basidiomycota, Polypores) from China Chen YY, Wu F* Institute of Microbiology, Beijing Forestry University, Beijing 100083, China Chen YY, Wu F 2017 –A new species of Antrodia (Basidiomycota, Polypores) from China. Mycosphere 8(7), 878–885, Doi 10.5943/mycosphere/8/7/4 Abstract A new species, Antrodia monomitica sp. nov., is described and illustrated from China based on morphological characters and molecular evidence. It is characterized by producing annual, fragile and nodulose basidiomata, a monomitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled and fusiform to mango-shaped basidiospores (6–7.5 × 2.3– 3 µm), and causing a typical brown rot. In phylogenetic analysis inferred from ITS and nLSU rDNA sequences, the new species forms a distinct lineage in the Antrodia s. l., and has a close relationship with A. oleracea. Key words – Fomitopsidaceae – phylogenetic analysis – taxonomy – wood-decaying fungi Introduction Antrodia P. Karst., typified with Polyporus serpens Fr. (=Antrodia albida (Fr.) Donk (Donk 1960, Ryvarden 1991), is characterized by a resupinate to effused-reflexed growth habit, white or pale colour of the context, a dimitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled, cylindrical to very narrow ellipsoid basidiospores which are negative in Melzer’s reagent and Cotton Blue, and causing a brown rot (Ryvarden & Melo 2014). Antrodia is a highly heterogeneous genus which is closely related to Fomitopsis P. -

A Phylogenetic Overview of the Antrodia Clade (Basidiomycota, Polyporales)
Mycologia, 105(6), 2013, pp. 1391–1411. DOI: 10.3852/13-051 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales) Beatriz Ortiz-Santana1 phylogenetic studies also have recognized the genera Daniel L. Lindner Amylocystis, Dacryobolus, Melanoporia, Pycnoporellus, US Forest Service, Northern Research Station, Center for Sarcoporia and Wolfiporia as part of the antrodia clade Forest Mycology Research, One Gifford Pinchot Drive, (SY Kim and Jung 2000, 2001; Binder and Hibbett Madison, Wisconsin 53726 2002; Hibbett and Binder 2002; SY Kim et al. 2003; Otto Miettinen Binder et al. 2005), while the genera Antrodia, Botanical Museum, University of Helsinki, PO Box 7, Daedalea, Fomitopsis, Laetiporus and Sparassis have 00014, Helsinki, Finland received attention in regard to species delimitation (SY Kim et al. 2001, 2003; KM Kim et al. 2005, 2007; Alfredo Justo Desjardin et al. 2004; Wang et al. 2004; Wu et al. 2004; David S. Hibbett Dai et al. 2006; Blanco-Dios et al. 2006; Chiu 2007; Clark University, Biology Department, 950 Main Street, Worcester, Massachusetts 01610 Lindner and Banik 2008; Yu et al. 2010; Banik et al. 2010, 2012; Garcia-Sandoval et al. 2011; Lindner et al. 2011; Rajchenberg et al. 2011; Zhou and Wei 2012; Abstract: Phylogenetic relationships among mem- Bernicchia et al. 2012; Spirin et al. 2012, 2013). These bers of the antrodia clade were investigated with studies also established that some of the genera are molecular data from two nuclear ribosomal DNA not monophyletic and several modifications have regions, LSU and ITS. A total of 123 species been proposed: the segregation of Antrodia s.l. -

Enzymatic Plant Cell Wall Degradation by the White Rot Fungus Dichomitus Squalens
YEB Recent Publications in this Series JOHANNA RYTIOJA Enzymatic Plant Cell Wall Degradation by the White Rot Fungus 33/2015 Niina Idänheimo The Role of Cysteine-rich Receptor-like Protein Kinases in ROS signaling in Arabidopsis thaliana 34/2015 Susanna Keriö Terpene Analysis and Transcript Profiling of the Conifer Response to Heterobasidion annosum s.l. Infection and Hylobius abietis Feeding 35/2015 Ann-Katrin Llarena DISSERTATIONES SCHOLA DOCTORALIS SCIENTIAE CIRCUMIECTALIS, Population Genetics and Molecular Epidemiology of Campylobacter jejuni ALIMENTARIAE, BIOLOGICAE. UNIVERSITATIS HELSINKIENSIS 15/2016 1/2016 Hanna Help-Rinta-Rahko The Interaction Of Auxin and Cytokinin Signalling Regulates Primary Root Procambial Patterning, Xylem Cell Fate and Differentiation in Arabidopsis thaliana 2/2016 Abbot O. Oghenekaro Molecular Analysis of the Interaction between White Rot Pathogen (Rigidoporus microporus) and Rubber Tree (Hevea brasiliensis) JOHANNA RYTIOJA 3/2016 Stiina Rasimus-Sahari Effects of Microbial Mitochondriotoxins from Food and Indoor Air on Mammalian Cells 4/2016 Hany S.M. EL Sayed Bashandy Enzymatic Plant Cell Wall Degradation by the Flavonoid Metabolomics in Gerbera hybrida and Elucidation of Complexity in the Flavonoid Biosynthetic Pathway White Rot Fungus Dichomitus squalens 5/2016 Erja Koivunen Home-Grown Grain Legumes in Poultry Diets 6/2016 Paul Mathijssen Holocene Carbon Dynamics and Atmospheric Radiative Forcing of Different Types of Peatlands in Finland 7/2016 Seyed Abdollah Mousavi Revised Taxonomy of the Family