Predicting Species Distributions from Small Numbers of Occurrence Records
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PRAVILNIK O PREKOGRANIĈNOM PROMETU I TRGOVINI ZAŠTIĆENIM VRSTAMA ("Sl
PRAVILNIK O PREKOGRANIĈNOM PROMETU I TRGOVINI ZAŠTIĆENIM VRSTAMA ("Sl. glasnik RS", br. 99/2009 i 6/2014) I OSNOVNE ODREDBE Ĉlan 1 Ovim pravilnikom propisuju se: uslovi pod kojima se obavlja uvoz, izvoz, unos, iznos ili tranzit, trgovina i uzgoj ugroţenih i zaštićenih biljnih i ţivotinjskih divljih vrsta (u daljem tekstu: zaštićene vrste), njihovih delova i derivata; izdavanje dozvola i drugih akata (potvrde, sertifikati, mišljenja); dokumentacija koja se podnosi uz zahtev za izdavanje dozvola, sadrţina i izgled dozvole; spiskovi vrsta, njihovih delova i derivata koji podleţu izdavanju dozvola, odnosno drugih akata; vrste, njihovi delovi i derivati ĉiji je uvoz odnosno izvoz zabranjen, ograniĉen ili obustavljen; izuzeci od izdavanja dozvole; naĉin obeleţavanja ţivotinja ili pošiljki; naĉin sprovoĊenja nadzora i voĊenja evidencije i izrada izveštaja. Ĉlan 2 Izrazi upotrebljeni u ovom pravilniku imaju sledeće znaĉenje: 1) datum sticanja je datum kada je primerak uzet iz prirode, roĊen u zatoĉeništvu ili veštaĉki razmnoţen, ili ukoliko takav datum ne moţe biti dokazan, sledeći datum kojim se dokazuje prvo posedovanje primeraka; 2) deo je svaki deo ţivotinje, biljke ili gljive, nezavisno od toga da li je u sveţem, sirovom, osušenom ili preraĊenom stanju; 3) derivat je svaki preraĊeni deo ţivotinje, biljke, gljive ili telesna teĉnost. Derivati većinom nisu prepoznatljivi deo primerka od kojeg potiĉu; 4) država porekla je drţava u kojoj je primerak uzet iz prirode, roĊen i uzgojen u zatoĉeništvu ili veštaĉki razmnoţen; 5) druga generacija potomaka -

Cop13 Analyses Cover 29 Jul 04.Qxd
IUCN/TRAFFIC Analyses of the Proposals to Amend the CITES Appendices at the 13th Meeting of the Conference of the Parties Bangkok, Thailand 2-14 October 2004 Prepared by IUCN Species Survival Commission and TRAFFIC Production of the 2004 IUCN/TRAFFIC Analyses of the Proposals to Amend the CITES Appendices was made possible through the support of: The Commission of the European Union Canadian Wildlife Service Ministry of Agriculture, Nature and Food Quality, Department for Nature, the Netherlands Federal Agency for Nature Conservation, Germany Federal Veterinary Office, Switzerland Ministerio de Medio Ambiente, Dirección General para la Biodiversidad (Spain) Ministère de l'écologie et du développement durable, Direction de la nature et des paysages (France) IUCN-The World Conservation Union IUCN-The World Conservation Union brings together states, government agencies and a diverse range of non-governmental organizations in a unique global partnership - over 1 000 members in some 140 countries. As a Union, IUCN seeks to influence, encourage and assist societies throughout the world to conserve the integrity and diversity of nature and to ensure that any use of natural resources is equitable and ecologically sustainable. IUCN builds on the strengths of its members, networks and partners to enhance their capacity and to support global alliances to safeguard natural resources at local, regional and global levels. The Species Survival Commission (SSC) is the largest of IUCN’s six volunteer commissions. With 8 000 scientists, field researchers, government officials and conservation leaders, the SSC membership is an unmatched source of information about biodiversity conservation. SSC members provide technical and scientific advice to conservation activities throughout the world and to governments, international conventions and conservation organizations. -

No 811/2008 of 13 August 2008 Suspending the Introduction Into the Community of Specimens of Certain Species of Wild Fauna and Flora
14.8.2008EN Official Journal of the European Union L 219/17 COMMISSION REGULATION (EC) No 811/2008 of 13 August 2008 suspending the introduction into the Community of specimens of certain species of wild fauna and flora THE COMMISSION OF THE EUROPEAN COMMUNITIES, — Accipiter erythropus, Aquila rapax, Gyps africanus, Lophaetus occipitalis and Poicephalus gulielmi from Guinea, Having regard to the Treaty establishing the European Community, — Hieraaetus ayresii, Hieraaetus spilogaster, Polemaetus bellicosus, Falco chicquera, Varanus ornatus (wild and Having regard to Council Regulation (EC) No 338/97 of ranched specimens) and Calabaria reinhardtii (wild 9 December 1996 on the protection of species of wild fauna specimens) from Togo, and flora by regulating trade therein (1), and in particular Article 19(2) thereof, — Agapornis pullarius and Poicephalus robustus from Côte d’Ivoire, After consulting the Scientific Review Group, — Stephanoaetus coronatus from Côte d’Ivoire and Togo, Whereas: — Pyrrhura caeruleiceps from Colombia; Pyrrhura pfrimeri (1) Article 4(6) of Regulation (EC) No 338/97 provides that from Brazil, the Commission may establish restrictions to the intro duction of certain species into the Community in accordance with the conditions laid down in points (a) — Brookesia decaryi, Uroplatus ebenaui, Uroplatus fimbriatus, to (d) thereof. Furthermore, implementing measures for Uroplatus guentheri, Uroplatus henkeli, Uroplatus lineatus, such restrictions have been laid down in Commission Uroplatus malama, Uroplatus phantasticus, Uroplatus Regulation (EC) No 865/2006 of 4 May 2006 laying pietschmanni, Uroplatus sikorae, Euphorbia ankarensis, down detailed rules concerning the implementation of Euphorbia berorohae, Euphorbia bongolavensis, Euphorbia Council Regulation (EC) No 338/97 of the protection duranii, Euphorbia fiananantsoae, Euphorbia iharanae, of species of wild fauna and flora by regulating trade Euphorbia labatii, Euphorbia lophogona, Euphorbia 2 therein ( ). -
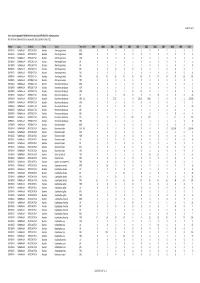
Gross Trade in Appendix II FAUNA (Direct Trade Only), 1999-2010 (For
AC25 Inf. 5 (1) Gross trade in Appendix II FAUNA (direct trade only), 1999‐2010 (for selection process) N.B. Data from 2009 and 2010 are incomplete. Data extracted 1 April 2011 Phylum Class TaxOrder Family Taxon Term Unit 1999 2000 2001 2002 2003 2004 2005 2006 2007 2008 2009 Total CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia BOD 0 00001000102 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia BON 0 00080000008 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia HOR 0 00000110406 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia LIV 0 00060000006 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia SKI 1 11311000008 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia SKP 0 00000010001 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia SKU 2 052101000011 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia TRO 15 42 49 43 46 46 27 27 14 37 26 372 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Antilope cervicapra TRO 0 00000020002 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae BOD 0 00100001002 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae HOP 0 00200000002 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae HOR 0 0010100120216 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae LIV 0 0 5 14 0 0 0 30 0 0 0 49 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae MEA KIL 0 5 27.22 0 0 272.16 1000 00001304.38 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae MEA 0 00000000101 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae -

COMMISSION IMPLEMENTING REGULATION (EU) 2017/1915 Of
20.10.2017 EN Official Journal of the European Union L 271/7 COMMISSION IMPLEMENTING REGULATION (EU) 2017/1915 of 19 October 2017 prohibiting the introduction into the Union of specimens of certain species of wild fauna and flora THE EUROPEAN COMMISSION, Having regard to the Treaty on the Functioning of the European Union, Having regard to Council Regulation (EC) No 338/97 of 9 December 1996 on the protection of species of wild fauna and flora by regulating trade therein (1), and in particular Article 4(6) thereof, Whereas: (1) The purpose of Regulation (EC) No 338/97 is to protect species of wild fauna and flora and to guarantee their conservation by regulating trade in animal and plant species listed in its Annexes. The species listed in the Annexes include the species set out in the Appendices to the Convention on International Trade in Endangered Species of Wild Fauna and Flora signed in 1973 (2) (the Convention) as well as species whose conservation status requires that trade from, into and within the Union be regulated or monitored. (2) Article 4(6) of Regulation (EC) No 338/97 provides that the Commission may establish restrictions to the introduction of specimens of certain species into the Union in accordance with the conditions laid down in points (a) to (d) thereof. (3) On the basis of recent information, the Scientific Review Group established pursuant to Article 17 of Regulation (EC) No 338/97 has concluded that the conservation status of certain species listed in Annex B to Regulation (EC) No 338/97 would be seriously jeopardised if their introduction into the Union from certain countries of origin is not prohibited. -

Biologie, Ecologie Et Conservation Animales ETUDE ECO-BIOLOGIQUES DES UROPLATE
UNIVERSITE D’ANTANANARIVO FACULTE DES SCIENCES DEPARTEMENT DE BIOLOGIE ANIMALE DEPARTEMENT DE BIOLOGIE ANIMALE Latimeria chalumnae MEMOIRE POUR L’OBTENTION DU Diplôme d’Etudes Approfondies (D.E.A.) Formation Doctorale : Sciences de la vie Option : Biologie, Ecologie et Conservation Animales ETUDE ECO-BIOLOGIQUES DES UROPLATES ET IMPLICATION DES ANALYSES PHYLOGENETIQUES DANS LEUR SYSTEMATIQUE Présenté par : Melle RATSOAVINA Mihaja Fanomezana Devant le JURY composé de : Président : Madame RAMILIJAONA RAVOAHANGIMALALA Olga, Professeur Titulaire Rapporteurs : Madame RAZAFIMAHATRATRA Emilienne, Maître de Conférences Examinateurs: Madame RAZAFINDRAIBE Hanta, Maître de conférences Monsieur EDWARD LOUIS, Doctor of Veterinary Medicine, PhD Soutenu publiquement le 27 Février 2009 ________________________________________________________________Remerciements REMERCIEMENTS Cette étude a été réalisée dans le cadre de l’accord de collaboration entre le Département de Biologie Animale, Faculté des Sciences, Université d’Antananarivo et Henry Doorly Zoo Omaha. Nebraska. USA, dans le cadre du projet Madagascar Biogeography Project (MBP). Je tiens à exprimer ma profonde reconnaissance à: - Madame RAMILIJAONA RAVOAHANGIMALALA Olga, Professeur Titulaire, Responsable de la formation doctorale, d'avoir accepté de présider ce mémoire et consacré son précieux temps pour la commission de lecture. - Madame RAZAFIMAHATRATRA Emilienne, Maître de conférences, encadreur et rapporteur qui a accepté de diriger et de corriger ce mémoire. Je la remercie pour ses nombreux conseils. - Madame RAZAFINDRAIBE Hanta, Maître de conférences, qui a accepté de faire partie de la commission de lecture, de juger ce travail et de figurer parmi les membres du jury. - Monsieur Edward LOUIS, Doctor of Veterinary Medecine, PhD d’être parmi les membres du jury et qui m’a permis d’avoir intégrer l’équipe de HDZ et de m’avoir inculqué les a priori des travaux de recherche. -

AC27 Doc. 12.5
Original language: English AC27 Doc. 12.5 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________ Twenty-seventh meeting of the Animals Committee Veracruz (Mexico), 28 April – 3 May 2014 Interpretation and implementation of the Convention Review of Significant Trade in specimens of Appendix-II species [Resolution Conf. 12.8 (Rev. CoP13)] SELECTION OF SPECIES FOR TRADE REVIEWS FOLLOWING COP16 1. This document has been prepared by the Secretariat. 2. In Resolution Conf. 12.8 (Rev. CoP13) on Review of Significant Trade in specimens of Appendix-II species, the Conference of the Parties: DIRECTS the Animals and Plants Committees, in cooperation with the Secretariat and experts, and in consultation with range States, to review the biological, trade and other relevant information on Appendix-II species subject to significant levels of trade, to identify problems and solutions concerning the implementation of Article IV, paragraphs 2 (a), 3 and 6 (a)... 3. In accordance with paragraph a) of that Resolution under the section Regarding conduct of the Review of Significant Trade, the Secretariat requested UNEP-WCMC to produce a summary from the CITES Trade Database of annual report statistics showing the recorded net level of exports for Appendix-II species over the five most recent years. Its report is attached as Annex 1 (English only) to the present document. The raw data used to prepare this summary are available in document AC27 Inf. 2. 4. Paragraph b) of the same section directs the Animals Committee, on the basis of recorded trade levels and information available to it, the Secretariat, Parties or other relevant experts, to select species of priority concern for review (whether or not such species have been the subject of a previous review). -

Inventaire Faune Au Sein De La Nouvelle Aire Protegee De Beampingaratsy
RAPPORT FINAL Projet : INVENTAIRE FAUNE AU SEIN DE LA NOUVELLE AIRE PROTEGEE DE BEAMPINGARATSY - Contrat No. : TLK/NTD/20201001/001 - Début des travaux : 1er octobre 2020 - Fin des travaux : 31 mars 2021 Préparé pour NITIDÆ – Projet TALAKY Lot VE 26 L Ambanidia Antananarivo 101 Distribution • 1 Copie : NITIDÆ – Projet TALAKY • 1 Copie : ECOFAUNA Préparé par l’équipe EcoFauna : Dr Zafimahery RAKOTOMALALA Rijaniaina Jean Nary ANDRIANJAKA Sylvain Rija RAKOTOSOA Angelo François ANDRIANIAINA Jary Harinarivo RAZAFINDRAIBE TABLE DES MATIERES LISTE DES TABLEAUX ............................................................................................................. iv LISTE DES FIGURES ................................................................................................................ vi LISTES DE S ANNEXES ........................................................................................................... vii LISTE DES ACRONYMES ........................................................................................................ vii INTRODUCTION ......................................................................................................................... 1 METHODES D’INVENTAIRE BIOLOGIQUE ..................................................................... 3 I.1. Méthodes d’inventaire des Amphibiens et Reptiles .................................................... 3 I.1.1. Observation directe le long d’un itinéraire échantillon (Transect) ........................ 3 I.1.2. Capture relâche par trou-piège ou -

A Phylogeny and Revised Classification of Squamata, Including 4161 Species of Lizards and Snakes
BMC Evolutionary Biology This Provisional PDF corresponds to the article as it appeared upon acceptance. Fully formatted PDF and full text (HTML) versions will be made available soon. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes BMC Evolutionary Biology 2013, 13:93 doi:10.1186/1471-2148-13-93 Robert Alexander Pyron ([email protected]) Frank T Burbrink ([email protected]) John J Wiens ([email protected]) ISSN 1471-2148 Article type Research article Submission date 30 January 2013 Acceptance date 19 March 2013 Publication date 29 April 2013 Article URL http://www.biomedcentral.com/1471-2148/13/93 Like all articles in BMC journals, this peer-reviewed article can be downloaded, printed and distributed freely for any purposes (see copyright notice below). Articles in BMC journals are listed in PubMed and archived at PubMed Central. For information about publishing your research in BMC journals or any BioMed Central journal, go to http://www.biomedcentral.com/info/authors/ © 2013 Pyron et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes Robert Alexander Pyron 1* * Corresponding author Email: [email protected] Frank T Burbrink 2,3 Email: [email protected] John J Wiens 4 Email: [email protected] 1 Department of Biological Sciences, The George Washington University, 2023 G St. -

Continental Speciation in the Tropics: Contrasting Biogeographic Patterns of Divergence in the Uroplatus Leaf-Tailed Gecko Radiation of Madagascar C
Journal of Zoology Journal of Zoology. Print ISSN 0952-8369 Continental speciation in the tropics: contrasting biogeographic patterns of divergence in the Uroplatus leaf-tailed gecko radiation of Madagascar C. J. Raxworthy1, R. G. Pearson1, B. M. Zimkus1,Ã, S. Reddy1,w, A. J. Deo1,2,z, R. A. Nussbaum3 & C. M. Ingram1 1 American Museum of Natural History, New York, NY, USA 2 Department of Biology, New York University, New York, NY, USA 3 Division of Amphibians and Reptiles, Museum of Zoology, University of Michigan, Ann Arbor, MI, USA Keywords Abstract speciation; biogeography; systematics; Reptilia; Gekkonidae; Madagascar. A fundamental expectation of vicariance biogeography is for contemporary cladogenesis to produce spatial congruence between speciating sympatric clades. Correspondence The Uroplatus leaf-tailed geckos represent one of most spectacular reptile radia- Christopher J. Raxworthy, American tions endemic to the continental island of Madagascar, and thus serve as an Museum of Natural History, Central Park excellent group for examining patterns of continental speciation within this large West at 79th Street, New York, NY 10024- and comparatively isolated tropical system. Here we present the first phylogeny 5192, USA. Email: [email protected] that includes complete taxonomic sampling for the group, and is based on morphology and molecular (mitochondrial and nuclear DNA) data. This study ÃCurrent address: Department of includes all described species, and we also include data for eight new species. We Herpetology, Museum of Comparative find novel outgroup relationships for Uroplatus and find strongest support for Zoology, Harvard University, 26 Oxford Paroedura as its sister taxon. Uroplatus is estimated to have initially diverged Street, Cambridge, MA 02138, during the mid-Tertiary in Madagascar, and includes two major speciose radia- w USA. -

Review of Selected Species Subject to Long- Standing Import Suspensions
UNEP-WCMC technical report Review of selected species subject to long- standing import suspensions Part I: Africa (Version edited for public release) Review of selected species subject to long-standing import 2 suspensions. Part I: Africa Prepared for The European Commission, Directorate General Environment, Directorate E - Global & Regional Challenges, LIFE ENV.E.2. – Global Sustainability, Trade & Multilateral Agreements, Brussels, Belgium Prepared August 2015 Copyright European Commission 2015 Citation UNEP-WCMC. 2015. Review of selected species subject to long-standing import suspensions. Part I: Africa. UNEP-WCMC, Cambridge. The UNEP World Conservation Monitoring Centre (UNEP-WCMC) is the specialist biodiversity assessment of the United Nations Environment Programme, the world’s foremost intergovernmental environmental organization. The Centre has been in operation for over 30 years, combining scientific research with policy advice and the development of decision tools. We are able to provide objective, scientifically rigorous products and services to help decision- makers recognize the value of biodiversity and apply this knowledge to all that they do. To do this, we collate and verify data on biodiversity and ecosystem services that we analyze and interpret in comprehensive assessments, making the results available in appropriate forms for national and international level decision-makers and businesses. To ensure that our work is both sustainable and equitable we seek to build the capacity of partners where needed, so that they can provide the same services at national and regional scales. The contents of this report do not necessarily reflect the views or policies of UNEP, contributory organisations or editors. The designations employed and the presentations do not imply the expressions of any opinion whatsoever on the part of UNEP, the European Commission or contributory organisations, editors or publishers concerning the legal status of any country, territory, city area or its authorities, or concerning the delimitation of its frontiers or boundaries. -

Tetrapods on the EDGE: Overcoming Data Limitations to Identify Phylogenetic
bioRxiv preprint doi: https://doi.org/10.1101/232991; this version posted December 21, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Tetrapods on the EDGE: Overcoming data limitations to identify phylogenetic 2 conservation priorities 3 4 Rikki Gumbs1,2*, Claudia L. Gray2, Oliver R. Wearn2,3, Nisha R. Owen2 5 6 1 Science and Solutions for a Changing Planet DTP, and the Department of Life Sciences, Imperial 7 College London, Silwood Park Campus, Ascot, Berkshire, United Kingdom 8 2 EDGE of Existence Programme, Zoological Society of London, Regent’s Park, London 9 3Institute of Zoology, Zoological Society of London, Regent’s Park, London 10 11 * Corresponding author 12 E-mail: [email protected] (RG) 1 bioRxiv preprint doi: https://doi.org/10.1101/232991; this version posted December 21, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 13 Abstract 14 The scale of the ongoing biodiversity crisis requires both effective conservation prioritisation and urgent 15 action. The EDGE metric, which prioritises species based on their Evolutionary Distinctiveness (ED) 16 and Global Endangerment (GE), relies on adequate phylogenetic and extinction risk data to generate 17 meaningful priorities for conservation. However, comprehensive phylogenetic analyses of large clades 18 are extremely rare and, even when available, become quickly out-of-date due to the rapid rate of species 19 descriptions and taxonomic revisions.