Shoot Flammability of Vascular Plants Is Phylogenetically Conserved And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Full of Beans: a Study on the Alignment of Two Flowering Plants Classification Systems
Full of beans: a study on the alignment of two flowering plants classification systems Yi-Yun Cheng and Bertram Ludäscher School of Information Sciences, University of Illinois at Urbana-Champaign, USA {yiyunyc2,ludaesch}@illinois.edu Abstract. Advancements in technologies such as DNA analysis have given rise to new ways in organizing organisms in biodiversity classification systems. In this paper, we examine the feasibility of aligning two classification systems for flowering plants using a logic-based, Region Connection Calculus (RCC-5) ap- proach. The older “Cronquist system” (1981) classifies plants using their mor- phological features, while the more recent Angiosperm Phylogeny Group IV (APG IV) (2016) system classifies based on many new methods including ge- nome-level analysis. In our approach, we align pairwise concepts X and Y from two taxonomies using five basic set relations: congruence (X=Y), inclusion (X>Y), inverse inclusion (X<Y), overlap (X><Y), and disjointness (X!Y). With some of the RCC-5 relationships among the Fabaceae family (beans family) and the Sapindaceae family (maple family) uncertain, we anticipate that the merging of the two classification systems will lead to numerous merged solutions, so- called possible worlds. Our research demonstrates how logic-based alignment with ambiguities can lead to multiple merged solutions, which would not have been feasible when aligning taxonomies, classifications, or other knowledge or- ganization systems (KOS) manually. We believe that this work can introduce a novel approach for aligning KOS, where merged possible worlds can serve as a minimum viable product for engaging domain experts in the loop. Keywords: taxonomy alignment, KOS alignment, interoperability 1 Introduction With the advent of large-scale technologies and datasets, it has become increasingly difficult to organize information using a stable unitary classification scheme over time. -

Phylogeny of Rosids! ! Rosids! !
Phylogeny of Rosids! Rosids! ! ! ! ! Eurosids I Eurosids II Vitaceae Saxifragales Eurosids I:! Eurosids II:! Zygophyllales! Brassicales! Celastrales! Malvales! Malpighiales! Sapindales! Oxalidales! Myrtales! Fabales! Geraniales! Rosales! Cucurbitales! Fagales! After Jansen et al., 2007, Proc. Natl. Acad. Sci. USA 104: 19369-19374! Phylogeny of Rosids! Rosids! ! ! ! ! Eurosids I Eurosids II Vitaceae Saxifragales Eurosids I:! Eurosids II:! Zygophyllales! Brassicales! Celastrales! Malvales! Malpighiales! Sapindales! Oxalidales! Myrtales! Fabales! Geraniales! Rosales! Cucurbitales! Fagales! After Jansen et al., 2007, Proc. Natl. Acad. Sci. USA 104: 19369-19374! Alnus - alders A. rubra A. rhombifolia A. incana ssp. tenuifolia Alnus - alders Nitrogen fixation - symbiotic with the nitrogen fixing bacteria Frankia Alnus rubra - red alder Alnus rhombifolia - white alder Alnus incana ssp. tenuifolia - thinleaf alder Corylus cornuta - beaked hazel Carpinus caroliniana - American hornbeam Ostrya virginiana - eastern hophornbeam Phylogeny of Rosids! Rosids! ! ! ! ! Eurosids I Eurosids II Vitaceae Saxifragales Eurosids I:! Eurosids II:! Zygophyllales! Brassicales! Celastrales! Malvales! Malpighiales! Sapindales! Oxalidales! Myrtales! Fabales! Geraniales! Rosales! Cucurbitales! Fagales! After Jansen et al., 2007, Proc. Natl. Acad. Sci. USA 104: 19369-19374! Fagaceae (Beech or Oak family) ! Fagaceae - 9 genera/900 species.! Trees or shrubs, mostly northern hemisphere, temperate region ! Leaves simple, alternate; often lobed, entire or serrate, deciduous -
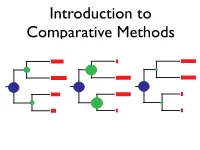
BM CC EB What Can We Learn from a Tree?
Introduction to Comparative Methods BM CC EB What can we learn from a tree? Net diversification (r) Relative extinction (ε) Peridiscaceae Peridiscaceae yllaceae yllaceae h h atop atop Proteaceae Proteaceae r r Ce Ce Tr oc T ho r M M o de c y y H H C C h r r e e a D D a o o nd o e e G G a a m m t t a a d r P r P h h e e u u c c p A p A r e a a e e a a a c a c n n a i B i B h h n d m d l m a l a m m e a e a e e t t n n c u u n n i i d i i e e e e o n o n p n p n a e a S e e S e e n n x x i i r c c a a n o n o p p h g e h g ae e l r a l r a a a a a a i i a a a e a e i b i b h y d c h d c i y i c a a c x c x c c G I a G I a n c n c c c y l y l t a a t a a e e e e e i l c i l c m l m l e c e c f a e a a f a e a a l r r l c c a i i r l e t e t a a r l a a e e u u u u o a o a a a c a c a a l a l e e e b b a a a a e e c e e c a a s c s c c e l c e l e e g e g e a a a a e e n n s e e s e e e e a a a P a P e e N N u u u S u S a e a e a a e e c c l n a l n e e a e e a e a e a e a e r a r a c c C i C i R R a e a e a e a e r c r c A A a d a a d a e i e i phanopetalaceae s r e ph s r e a a s e c s e c e e u u b a a b a e e P P r r l l e n e a a a a m m entho e e e Ha a H o a c r e c r e nt B B e p e e e c e e c a c c h e a p a a p a lo lo l l a a e s o t e s i a r a i a r r r r r a n e a n e a b a l b t a t gaceae e g e ceae a c a s c s a z e z M i a e M i a c a d e a d e ae e ae r e r a e a e a a a c c ce r e r L L i i ac a Vitaceae Vi r r C C e e ta v e v e a a c a a e ea p e ap c a c a e a P P e e l l e Ge G e e ae a t t e e p p r r ce c an u an -

Field Identification of the 50 Most Common Plant Families in Temperate Regions
Field identification of the 50 most common plant families in temperate regions (including agricultural, horticultural, and wild species) by Lena Struwe [email protected] © 2016, All rights reserved. Note: Listed characteristics are the most common characteristics; there might be exceptions in rare or tropical species. This compendium is available for free download without cost for non- commercial uses at http://www.rci.rutgers.edu/~struwe/. The author welcomes updates and corrections. 1 Overall phylogeny – living land plants Bryophytes Mosses, liverworts, hornworts Lycophytes Clubmosses, etc. Ferns and Fern Allies Ferns, horsetails, moonworts, etc. Gymnosperms Conifers, pines, cycads and cedars, etc. Magnoliids Monocots Fabids Ranunculales Rosids Malvids Caryophyllales Ericales Lamiids The treatment for flowering plants follows the APG IV (2016) Campanulids classification. Not all branches are shown. © Lena Struwe 2016, All rights reserved. 2 Included families (alphabetical list): Amaranthaceae Geraniaceae Amaryllidaceae Iridaceae Anacardiaceae Juglandaceae Apiaceae Juncaceae Apocynaceae Lamiaceae Araceae Lauraceae Araliaceae Liliaceae Asphodelaceae Magnoliaceae Asteraceae Malvaceae Betulaceae Moraceae Boraginaceae Myrtaceae Brassicaceae Oleaceae Bromeliaceae Orchidaceae Cactaceae Orobanchaceae Campanulaceae Pinaceae Caprifoliaceae Plantaginaceae Caryophyllaceae Poaceae Convolvulaceae Polygonaceae Cucurbitaceae Ranunculaceae Cupressaceae Rosaceae Cyperaceae Rubiaceae Equisetaceae Rutaceae Ericaceae Salicaceae Euphorbiaceae Scrophulariaceae -

An All-Taxa Biodiversity Inventory of the Huron Mountain Club
AN ALL-TAXA BIODIVERSITY INVENTORY OF THE HURON MOUNTAIN CLUB Version: August 2016 Cite as: Woods, K.D. (Compiler). 2016. An all-taxa biodiversity inventory of the Huron Mountain Club. Version August 2016. Occasional papers of the Huron Mountain Wildlife Foundation, No. 5. [http://www.hmwf.org/species_list.php] Introduction and general compilation by: Kerry D. Woods Natural Sciences Bennington College Bennington VT 05201 Kingdom Fungi compiled by: Dana L. Richter School of Forest Resources and Environmental Science Michigan Technological University Houghton, MI 49931 DEDICATION This project is dedicated to Dr. William R. Manierre, who is responsible, directly and indirectly, for documenting a large proportion of the taxa listed here. Table of Contents INTRODUCTION 5 SOURCES 7 DOMAIN BACTERIA 11 KINGDOM MONERA 11 DOMAIN EUCARYA 13 KINGDOM EUGLENOZOA 13 KINGDOM RHODOPHYTA 13 KINGDOM DINOFLAGELLATA 14 KINGDOM XANTHOPHYTA 15 KINGDOM CHRYSOPHYTA 15 KINGDOM CHROMISTA 16 KINGDOM VIRIDAEPLANTAE 17 Phylum CHLOROPHYTA 18 Phylum BRYOPHYTA 20 Phylum MARCHANTIOPHYTA 27 Phylum ANTHOCEROTOPHYTA 29 Phylum LYCOPODIOPHYTA 30 Phylum EQUISETOPHYTA 31 Phylum POLYPODIOPHYTA 31 Phylum PINOPHYTA 32 Phylum MAGNOLIOPHYTA 32 Class Magnoliopsida 32 Class Liliopsida 44 KINGDOM FUNGI 50 Phylum DEUTEROMYCOTA 50 Phylum CHYTRIDIOMYCOTA 51 Phylum ZYGOMYCOTA 52 Phylum ASCOMYCOTA 52 Phylum BASIDIOMYCOTA 53 LICHENS 68 KINGDOM ANIMALIA 75 Phylum ANNELIDA 76 Phylum MOLLUSCA 77 Phylum ARTHROPODA 79 Class Insecta 80 Order Ephemeroptera 81 Order Odonata 83 Order Orthoptera 85 Order Coleoptera 88 Order Hymenoptera 96 Class Arachnida 110 Phylum CHORDATA 111 Class Actinopterygii 112 Class Amphibia 114 Class Reptilia 115 Class Aves 115 Class Mammalia 121 INTRODUCTION No complete species inventory exists for any area. -

Botánica Sistemática II Guia De Actividades 2014
Facultad de Ciencias Naturales y Museo – Universidad Nacional de La Plata Cátedra de Botánica Sistematica II Botánica Sistemática II Guia de actividades 2014 1 Facultad de Ciencias Naturales y Museo – Universidad Nacional de La Plata Cátedra de Botánica Sistematica II Facultad de Ciencias Naturales y Museo Universidad Nacional de La Plata Cátedra de Botánica Sistemática II Profesor Titular: Dra. Susana E. Freire Jefes de Trabajos Prácticos: Lic. Gustavo Delucchi Lic. Laura Iharlegui Ayudantes: Lic. Marcelo Hernández Lic. Elena Rastelli Dra. Estrella Urtubey Lic. Carlos A. Zavaro Lic. Jessica Viera Barreto Lic. Damián Fernández Fernando Buet Pablo Simón La Plata, Abril 2014 2 Facultad de Ciencias Naturales y Museo – Universidad Nacional de La Plata Cátedra de Botánica Sistematica II Indice Actividades de las Clases Teórico-Prácticas Diversidad biológica. Crisis de la biodiversidad. El rol del sistemático………………………………...…6 Sistemas de Clasificación. Nomenclatura Botánica …………………………….…………………………...….…7 Concepto de Especie-Evidencia taxonómica: caracteres morfológicos y moleculares …….….…9 Evolución y especiación como explicación de la diversidad orgánica. …………………………………..10 Fuentes de información botánica ……………………………………………………………………………………….…11 Origen y características de las plantas terrestres (=Embriófitas). Briófitas. Origen de plantas vasculares (=Traqueófitas)……………………………………………………………………………..…………………12 Primeras plantas vasculares. Clases Rhyniopsida, Zosterophyllopsida, Trimerophytopsida, Subdivisión Lycopodiophytina: Clase Lycopodiopsida ..………………………………………………….…13 -

Characterization of the Complete Chloroplast Genome of Nitraria Tangutorum, a Desert Shrub
Journal of Genetics (2019) 98:91 © Indian Academy of Sciences https://doi.org/10.1007/s12041-019-1135-9 RESEARCH ARTICLE Characterization of the complete chloroplast genome of Nitraria tangutorum, a desert shrub MERHABA ABLA, XI ZHA, YING WANG, XIAO YANG WANG, FEI GAO∗ , YIJUN ZHOU and JINCHAO FENG College of Life and Environmental Sciences, Minzu University of China, Beijing 100081, People’s Republic of China *For correspondence. E-mail: [email protected]. Received 2 April 2019; revised 16 July 2019; accepted 19 July 2019; published online 5 September 2019 Abstract. The chloroplast genome sequence of Nitraria tangutorum, a desert shrub, was sequenced using high-throughput sequencing technology and analysed phylogenetically in the present study. The chloroplast genome is 159,414 bp in length, including a large single copy region of 87,924 bp and small single copy region of 18,318 bp, and a pair of inverted repeat regions of 26,586 bp. The chloroplast genome contains 110 unique genes, including 77 protein-coding genes, four ribosomal RNA genes, and 29 tRNA genes. Most of these genes are present as a single copy and in two or more copies 19 genes occurred. Seventeen genes have one intron, and clpP and ycf3 genes contain two introns. A total of 81 simple sequence repeats (SSRs) were identified, most of them were found to be mononucleotide repeats composed of A/T. In addition to SSRs, 66 repeats were identified, including 41 tandem repeats, 10 palindromic repeats, and 15 forward repeats. The phylogenetic analysis based on 54 protein-coding genes demonstrated a close relationship between N. -

Downloaded from Genbank on That Full Plastid Genomes Are Not Sufficient to Reject Al- February 28, 2012
Ruhfel et al. BMC Evolutionary Biology 2014, 14:23 http://www.biomedcentral.com/1471-2148/14/23 RESEARCH ARTICLE Open Access From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes Brad R Ruhfel1*, Matthew A Gitzendanner2,3,4, Pamela S Soltis3,4, Douglas E Soltis2,3,4 and J Gordon Burleigh2,4 Abstract Background: Next-generation sequencing has provided a wealth of plastid genome sequence data from an increasingly diverse set of green plants (Viridiplantae). Although these data have helped resolve the phylogeny of numerous clades (e.g., green algae, angiosperms, and gymnosperms), their utility for inferring relationships across all green plants is uncertain. Viridiplantae originated 700-1500 million years ago and may comprise as many as 500,000 species. This clade represents a major source of photosynthetic carbon and contains an immense diversity of life forms, including some of the smallest and largest eukaryotes. Here we explore the limits and challenges of inferring a comprehensive green plant phylogeny from available complete or nearly complete plastid genome sequence data. Results: We assembled protein-coding sequence data for 78 genes from 360 diverse green plant taxa with complete or nearly complete plastid genome sequences available from GenBank. Phylogenetic analyses of the plastid data recovered well-supported backbone relationships and strong support for relationships that were not observed in previous analyses of major subclades within Viridiplantae. However, there also is evidence of systematic error in some analyses. In several instances we obtained strongly supported but conflicting topologies from analyses of nucleotides versus amino acid characters, and the considerable variation in GC content among lineages and within single genomes affected the phylogenetic placement of several taxa. -

2 ANGIOSPERM PHYLOGENY GROUP (APG) SYSTEM History Of
ANGIOSPERM PHYLOGENY GROUP (APG) SYSTEM The Angiosperm Phylogeny Group, or APG, refers to an informal international group of systematic botanists who came together to try to establish a consensus view of the taxonomy of flowering plants (angiosperms) that would reflect new knowledge about their relationships based upon phylogenetic studies. As of 2010, three incremental versions of a classification system have resulted from this collaboration (published in 1998, 2003 and 2009). An important motivation for the group was what they viewed as deficiencies in prior angiosperm classifications, which were not based on monophyletic groups (i.e. groups consisting of all the descendants of a common ancestor). APG publications are increasingly influential, with a number of major herbaria changing the arrangement of their collections to match the latest APG system. Angiosperm classification and the APG Until detailed genetic evidence became available, the classification of flowering plants (also known as angiosperms, Angiospermae, Anthophyta or Magnoliophyta) was based on their morphology (particularly that of the flower) and their biochemistry (what kinds of chemical compound they contained or produced). Classification systems were typically produced by an individual botanist or by a small group. The result was a large number of such systems (see List of systems of plant taxonomy). Different systems and their updates tended to be favoured in different countries; e.g. the Engler system in continental Europe; the Bentham & Hooker system in Britain (particularly influential because it was used by Kew); the Takhtajan system in the former Soviet Union and countries within its sphere of influence; and the Cronquist system in the United States. -

Appendix 1. Uses and Destination of Species and Landraces Found in the Chagras
Appendix 1. Uses and destination of species and landraces found in the chagras. Chagra Order Family Genus Scientific name Landraces common Uses† Destination‡ names F M N T F SC S N Chagra 1 Poales BROMELIACEAE Ananas Ananas comosus Yellow peel pineapple x x Poales BROMELIACEAE Ananas Ananas comosus Red peel pineapple x x x Poales BROMELIACEAE Ananas Ananas comosus Green peel pineapple x x x Arecales ARECACEAE Euterpe Euterpe precatoria Asaí x x Solanales CONVOLVULACEAE Ipomoea Ipomoea batatas Camote x x Malpighiales EUPHORBIACEAE Manihot Manihot esculenta Flor manioc (sweet) x x x Malpighiales EUPHORBIACEAE Manihot Manihot esculenta Lupuna manioc (bitter) x x x Malpighiales EUPHORBIACEAE Manihot Manihot esculenta Mandioca manioc (bitter) x x x Malpighiales EUPHORBIACEAE Manihot Manihot esculenta Tresmesina manioc x x x (bitter) Malpighiales EUPHORBIACEAE Manihot Manihot esculenta Vega manioc (sweet) x x x Zingiberales MUSACEAE Musa Musa paradisiaca Banana x x x Zingiberales MUSACEAE Musa Musa sp Bellaco plantain x x x Zingiberales MUSACEAE Musa Musa sp Manzana plantain x x x Zingiberales MUSACEAE Musa Musa sp Píldoro/guineo plantain x x x Zingiberales MUSACEAE Musa Musa sp Propio/común plantain x x x Laurales LAURACEAE Persea Persea americana Avocado x x Poales POACEAE Saccharum Saccharum Sugar cane x x x officinarum Malvales MALVACEAE Theobroma Theobroma bicolor Macambo x x -

Ntegrated Pest Management of Longan (Sapindales: Sapindaceae) in Vietnam
Faculty Scholarship 2019 ntegrated Pest Management of Longan (Sapindales: Sapindaceae) in Vietnam Hanh Tran Hoa Nguyen Van Rangaswamy Muniappan James Amrine Rayapati Naidu See next page for additional authors Follow this and additional works at: https://researchrepository.wvu.edu/faculty_publications Part of the Plant Sciences Commons Authors Hanh Tran, Hoa Nguyen Van, Rangaswamy Muniappan, James Amrine, Rayapati Naidu, Robert Cilbertson, and Jaspreet Sidhu Journal of Integrated Pest Management, (2019) 10(1): 18; 1–10 doi: 10.1093/jipm/pmz016 Recommendations Integrated Pest Management of Longan (Sapindales: Sapindaceae) in Vietnam Hanh Tr a n ,1 Hoa Nguyen Van,1 Rangaswamy Muniappan,2,7 James Amrine,3 Rayapati Naidu,4 Robert Gilbertson,5 and Jaspreet Sidhu6 1 2 Plant Protection Division, Southern Horticultural Research Institute, Box 203, My Tho city, Tien Giang, Vietnam, Integrated Pest Downloaded from https://academic.oup.com/jipm/article-abstract/10/1/18/5510771 by guest on 28 April 2020 Management Innovation Lab, Virginia Tech, 526 Prices Fork Road, Blacksburg, VA 24061, 3Division of Plant and Soil Sciences, West Virginia University, Morgantown, WV 26506, 4Department of Plant Pathology, Irrigated Agriculture Research & Extension Center, Washington State University, Prosser, WA 99350, 5Department of Plant Pathology, University of California, Davis, CA 95616, 6Uni- versity of California Cooperative Extension, 1031 S Mount Vernon Ave, Bakersfield, CA 93307, and7 Corresponding author, e-mail: [email protected] Subject Editor: Tom Royer Received 17 January 2019; Editorial decision 29 April 2019 Abstract This paper describes the current state of pests and diseases of longan (Dimocarpus longan Lour.) and their management in Vietnam. Longan is the third most cultivated fruit crop and second major fruit crop exported from Vietnam. -

Renfrew County Plants
Renfrew County Plant Checklist. November 14, 2010. Scientific Name Common Name Author Kingdom Phylum Class Order Family Genus Species Subspecies Abies balsamea Balsam Fir (L.) P. Mill. Plantae Coniferophyta Pinopsida Pinales Pinaceae Abies balsamea Acalypha virginica Virginia Copperleaf L. Plantae Anthophyta Dicotyledoneae Euphorbiales Euphorbiaceae Acalypha virginica Acer negundo Box Elder L. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer negundo Acer nigrum Black Maple Michx. f. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer nigrum Acer pensylvanicum Striped Maple L. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer pensylvanicum Acer platanoides Norway Maple L. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer platanoides Acer rubrum Red Maple L. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer rubrum Acer saccharinum Silver Maple L. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer saccharinum Acer saccharum var. saccharum Sugar Maple Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer saccharum var. saccharum Acer spicatum Mountain Maple Lam. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer spicatum Acer x freemanii Hybrid Maple E. Murr. Plantae Anthophyta Dicotyledoneae Sapindales Aceraceae Acer x freemanii Achillea millefolium var. millefolium Common Yarrow Plantae Anthophyta Dicotyledoneae Asterales Asteraceae Achillea millefolium var. millefolium Achillea ptarmica False Sneezewort L. Plantae Anthophyta Dicotyledoneae Asterales Asteraceae Achillea