Identification of Clematidis Radix Et Rhizoma and Its Adulterants by Core Haplotype Based on the ITS Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Educated Youth Should Go to the Rural Areas: a Tale of Education, Employment and Social Values*
Educated Youth Should Go to the Rural Areas: A Tale of Education, Employment and Social Values* Yang You† Harvard University This draft: July 2018 Abstract I use a quasi-random urban-dweller allocation in rural areas during Mao’s Mass Rustication Movement to identify human capital externalities in education, employment, and social values. First, rural residents acquired an additional 0.1-0.2 years of education from a 1% increase in the density of sent-down youth measured by the number of sent-down youth in 1969 over the population size in 1982. Second, as economic outcomes, people educated during the rustication period suffered from less non-agricultural employment in 1990. Conversely, in 2000, they enjoyed increased hiring in all non-agricultural occupations and lower unemployment. Third, sent-down youth changed the social values of rural residents who reported higher levels of trust, enhanced subjective well-being, altered trust from traditional Chinese medicine to Western medicine, and shifted job attitudes from objective cognitive assessments to affective job satisfaction. To explore the mechanism, I document that sent-down youth served as rural teachers with two new county-level datasets. Keywords: Human Capital Externality, Sent-down Youth, Rural Educational Development, Employment Dynamics, Social Values, Culture JEL: A13, N95, O15, I31, I25, I26 * This paper was previously titled and circulated, “Does living near urban dwellers make you smarter” in 2017 and “The golden era of Chinese rural education: evidence from Mao’s Mass Rustication Movement 1968-1980” in 2015. I am grateful to Richard Freeman, Edward Glaeser, Claudia Goldin, Wei Huang, Lawrence Katz, Lingsheng Meng, Nathan Nunn, Min Ouyang, Andrei Shleifer, and participants at the Harvard Economic History Lunch Seminar, Harvard Development Economics Lunch Seminar, and Harvard China Economy Seminar, for their helpful comments. -

Economic Strategy for the Sustainable Development of Ice-Snow Tourism in Heilongjiang Province
The Frontiers of Society, Science and Technology ISSN 2616-7433 Vol. 1, Issue 5: 130-134, DOI: 10.25236/FSST.19010523 Economic Strategy for the Sustainable Development of Ice-snow Tourism in Heilongjiang Province Zhongquan Ma1, Fang Cao2 1. Harbin Institute of Technology, Heilongjiang 150001, China 2. Bohai University, Liaoning 121013, China ABSTRACT. Heilongjiang Province has unique ice-snow tourism resources, with the largest snow fall and the longest snow period in China. Its annual snow and ice period is 4 to 5 months. Hence, it is the province with the best ice-snow tourism conditions in China. Heilongjiang Province established Harbin Harbin Ice Lantern Exhibition in 1963, held the Ice and Snow Festival in 1985 and the Ice and Snow World in 1998. It has formed its own characteristics and advantages of ice-snow tourism and promoted the economic development of Heilongjiang Province. With the further development of science, technology and economy, the ice-snow tourism in Heilongjiang has entered a new stage of development. At this stage, it is an important issue worth studying that how to achieve sustainable development of ice- snow tourism in Heilongjiang Province. KEYWORDS: Heilongjiang province; Ice-snow tourism; Sustainable development; Economic strategy 1. Introduction Heilongjiang Province is the most northern province with the most latitude in China. The 0 ℃ contour of the annual average temperature passes through the middle part of the province. With the earliest snowfall and the latest snowfall in China, Heilongjiang Province has heavy snowfall and long snow period. The snow is clean and rich, with moderate hardness. It is the best province to develop ice-snow tourism in China. -

China Russia
1 1 1 1 Acheng 3 Lesozavodsk 3 4 4 0 Didao Jixi 5 0 5 Shuangcheng Shangzhi Link? ou ? ? ? ? Hengshan ? 5 SEA OF 5 4 4 Yushu Wuchang OKHOTSK Dehui Mudanjiang Shulan Dalnegorsk Nongan Hailin Jiutai Jishu CHINA Kavalerovo Jilin Jiaohe Changchun RUSSIA Dunhua Uglekamensk HOKKAIDOO Panshi Huadian Tumen Partizansk Sapporo Hunchun Vladivostok Liaoyuan Chaoyang Longjing Yanji Nahodka Meihekou Helong Hunjiang Najin Badaojiang Tong Hua Hyesan Kanggye Aomori Kimchaek AOMORI ? ? 0 AKITA 0 4 DEMOCRATIC PEOPLE'S 4 REPUBLIC OF KOREA Akita Morioka IWATE SEA O F Pyongyang GULF OF KOREA JAPAN Nampo YAMAJGATAA PAN Yamagata MIYAGI Sendai Haeju Niigata Euijeongbu Chuncheon Bucheon Seoul NIIGATA Weonju Incheon Anyang ISIKAWA ChechonREPUBLIC OF HUKUSIMA Suweon KOREA TOTIGI Cheonan Chungju Toyama Cheongju Kanazawa GUNMA IBARAKI TOYAMA PACIFIC OCEAN Nagano Mito Andong Maebashi Daejeon Fukui NAGANO Kunsan Daegu Pohang HUKUI SAITAMA Taegu YAMANASI TOOKYOO YELLOW Ulsan Tottori GIFU Tokyo Matsue Gifu Kofu Chiba SEA TOTTORI Kawasaki KANAGAWA Kwangju Masan KYOOTO Yokohama Pusan SIMANE Nagoya KANAGAWA TIBA ? HYOOGO Kyoto SIGA SIZUOKA ? 5 Suncheon Chinhae 5 3 Otsu AITI 3 OKAYAMA Kobe Nara Shizuoka Yeosu HIROSIMA Okayama Tsu KAGAWA HYOOGO Hiroshima OOSAKA Osaka MIE YAMAGUTI OOSAKA Yamaguchi Takamatsu WAKAYAMA NARA JAPAN Tokushima Wakayama TOKUSIMA Matsuyama National Capital Fukuoka HUKUOKA WAKAYAMA Jeju EHIME Provincial Capital Cheju Oita Kochi SAGA KOOTI City, town EAST CHINA Saga OOITA Major Airport SEA NAGASAKI Kumamoto Roads Nagasaki KUMAMOTO Railroad Lake MIYAZAKI River, lake JAPAN KAGOSIMA Miyazaki International Boundary Provincial Boundary Kagoshima 0 12.5 25 50 75 100 Kilometers Miles 0 10 20 40 60 80 ? ? ? ? 0 5 0 5 3 3 4 4 1 1 1 1 The boundaries and names show n and t he designations us ed on this map do not imply of ficial endors ement or acceptance by the United N at ions. -

Religion in China BKGA 85 Religion Inchina and Bernhard Scheid Edited by Max Deeg Major Concepts and Minority Positions MAX DEEG, BERNHARD SCHEID (EDS.)
Religions of foreign origin have shaped Chinese cultural history much stronger than generally assumed and continue to have impact on Chinese society in varying regional degrees. The essays collected in the present volume put a special emphasis on these “foreign” and less familiar aspects of Chinese religion. Apart from an introductory article on Daoism (the BKGA 85 BKGA Religion in China prototypical autochthonous religion of China), the volume reflects China’s encounter with religions of the so-called Western Regions, starting from the adoption of Indian Buddhism to early settlements of religious minorities from the Near East (Islam, Christianity, and Judaism) and the early modern debates between Confucians and Christian missionaries. Contemporary Major Concepts and religious minorities, their specific social problems, and their regional diversities are discussed in the cases of Abrahamitic traditions in China. The volume therefore contributes to our understanding of most recent and Minority Positions potentially violent religio-political phenomena such as, for instance, Islamist movements in the People’s Republic of China. Religion in China Religion ∙ Max DEEG is Professor of Buddhist Studies at the University of Cardiff. His research interests include in particular Buddhist narratives and their roles for the construction of identity in premodern Buddhist communities. Bernhard SCHEID is a senior research fellow at the Austrian Academy of Sciences. His research focuses on the history of Japanese religions and the interaction of Buddhism with local religions, in particular with Japanese Shintō. Max Deeg, Bernhard Scheid (eds.) Deeg, Max Bernhard ISBN 978-3-7001-7759-3 Edited by Max Deeg and Bernhard Scheid Printed and bound in the EU SBph 862 MAX DEEG, BERNHARD SCHEID (EDS.) RELIGION IN CHINA: MAJOR CONCEPTS AND MINORITY POSITIONS ÖSTERREICHISCHE AKADEMIE DER WISSENSCHAFTEN PHILOSOPHISCH-HISTORISCHE KLASSE SITZUNGSBERICHTE, 862. -

Investigation of Borrelia Spp. in Ticks (Acari: Ixodidae)
Asian Pacific Journal of Tropical Medicine (2012)459-464 459 Contents lists available at ScienceDirect Asian Pacific Journal of Tropical Medicine journal homepage:www.elsevier.com/locate/apjtm Document heading doi: Investigation of Borrelia spp. in ticks (Acari: Ixodidae) at the border crossings between China and Russia in Heilongjiang Province, China Shi Liu1, Chao Yuan2, Yun-Fu Cui1*, Bai-Xiang Li3, Li-Jie Wu3, Ying Liu4 1The 2nd Affiliated Hospital of Harbin Medical University, Harbin 150001, People s Republic of China 2 ' Daqing Oilfield General Hospital Group Rangbei Hospital, Daqing, 163114, People s Republic of China 3 ' Harbin Medical University School of Public Health, Harbin 150001, People s Republic of China 4 ' The 3rd Affiliated Hospital of Qiqihar Medical College, QiqiHar 161002, People s Republic of China ' ARTICLE INFO ABSTRACT Article history: Objective: Borrelia To investigate the precise species of tick vector andMethods: the spirochete pathogen Received 15 February 2012 at the Heilongjiang Province international border with Russia. In this study, ticks were Received in revised form 15 March 2012 collected from 12 Heilongjiang border crossings (including grasslands, shrublands, forests, and Accepted 15 April 2012 plantantions) to determine the rate and species type of spirochete-infected ticks and the most Available online 20 June 2012 Results: prevalent spirochete genotypes. The ticks represented three genera and four species Ixodes persulcatus Dermacentor silvarum Haemaphysalis concinna of the Ixodidae family [ , , and Haemaphysalis japonica Ixodes persulcatus Borrelia burgdorferi sensu ]. had the highest amount of Keywords: lato Borrelia Ixodes persulcatus infection of 25.6% and the most common species of isolated from Borrelia garinii Conclusions: Borrelia garinii Lyme disease was , strain PD91. -

Annual Report 2019
HAITONG SECURITIES CO., LTD. 海通證券股份有限公司 Annual Report 2019 2019 年度報告 2019 年度報告 Annual Report CONTENTS Section I DEFINITIONS AND MATERIAL RISK WARNINGS 4 Section II COMPANY PROFILE AND KEY FINANCIAL INDICATORS 8 Section III SUMMARY OF THE COMPANY’S BUSINESS 25 Section IV REPORT OF THE BOARD OF DIRECTORS 33 Section V SIGNIFICANT EVENTS 85 Section VI CHANGES IN ORDINARY SHARES AND PARTICULARS ABOUT SHAREHOLDERS 123 Section VII PREFERENCE SHARES 134 Section VIII DIRECTORS, SUPERVISORS, SENIOR MANAGEMENT AND EMPLOYEES 135 Section IX CORPORATE GOVERNANCE 191 Section X CORPORATE BONDS 233 Section XI FINANCIAL REPORT 242 Section XII DOCUMENTS AVAILABLE FOR INSPECTION 243 Section XIII INFORMATION DISCLOSURES OF SECURITIES COMPANY 244 IMPORTANT NOTICE The Board, the Supervisory Committee, Directors, Supervisors and senior management of the Company warrant the truthfulness, accuracy and completeness of contents of this annual report (the “Report”) and that there is no false representation, misleading statement contained herein or material omission from this Report, for which they will assume joint and several liabilities. This Report was considered and approved at the seventh meeting of the seventh session of the Board. All the Directors of the Company attended the Board meeting. None of the Directors or Supervisors has made any objection to this Report. Deloitte Touche Tohmatsu (Deloitte Touche Tohmatsu and Deloitte Touche Tohmatsu Certified Public Accountants LLP (Special General Partnership)) have audited the annual financial reports of the Company prepared in accordance with PRC GAAP and IFRS respectively, and issued a standard and unqualified audit report of the Company. All financial data in this Report are denominated in RMB unless otherwise indicated. -

World Bank Document
Procurement Plan for Liaoning Medium City Infrastructure Project (LMCIP) in China Project information: Public Disclosure Authorized Public Disclosure Authorized Country: China Borrower: The People’s Republic of China. Project Name: Liaoning Medium City Infrastructure Project (LMCIP) Loan/Credit No.: 4831-CHA Project ID: P099992 Project Implementing Agency (PIA): Liaoning Urban Construction and Renewal Project Office (LUCRPO) in Liaoning Province and city PMOs in cities of Fushun, Benxi, Liaoyang, Jinzhou, Panjin, and Dengta . Bank’s approval Date of the procurement Plan [Original:During Loan negotiation in May 2006; Revision 1:…] Date of General Procurement Notice: April 3, 2006 Period covered by this procurement plan: 2006-2009 Public Disclosure Authorized Public Disclosure Authorized The prior review thresholds for LMCI Project: Table A Civil Works Goods Consultant Consultant services services Firm Individual Above USD 5 million 500K 200K 50K In addition, the Bank will review the first contract procured under each category. The procurement method thresholds for LMCI Project : Public Disclosure Authorized Public Disclosure Authorized Table B Civil Goods Consultant services Works Public Disclosure Authorized Public Disclosure Authorized ICB >15 >500K >300K(short list not more than million 2 from a country) NCB advertisement >2 >300K <300K (shortlist can be only on a national million from national consultants) newspaper NCB advertisement <2 <300K >200K: QCBS on a provincial million <200K: CQS or Individual newspaper Consultant (IC) Shopping -

Characteristics of Spatial Connection Based on Intercity Passenger Traffic Flow in Harbin- Changchun Urban Agglomeration, China Research Paper
Guo, R.; Wu, T.; Wu, X.C. Characteristics of Spatial Connection Based on Intercity Passenger Traffic Flow in Harbin- Changchun Urban Agglomeration, China Research Paper Characteristics of Spatial Connection Based on Intercity Passenger Traffic Flow in Harbin-Changchun Urban Agglomeration, China Rong Guo, School of Architecture,Harbin Institute of Technology,Key Laboratory of Cold Region Urban and Rural Human Settlement Environment Science and Technology,Ministry of Industry and Information Technology,Harbin 150006,China Tong Wu, School of Architecture,Harbin Institute of Technology,Key Laboratory of Cold Region Urban and Rural Human Settlement Environment Science and Technology,Ministry of Industry and Information Technology,Harbin 150006,China Xiaochen Wu, School of Architecture,Harbin Institute of Technology,Key Laboratory of Cold Region Urban and Rural Human Settlement Environment Science and Technology,Ministry of Industry and Information Technology,Harbin 150006,China Abstract With the continuous improvement of transportation facilities and information networks, the obstruction of distance in geographic space has gradually weakened, and the hotspots of urban geography research have gradually changed from the previous city hierarchy to the characteristics of urban connections and networks. As the main carrier and manifestation of elements, mobility such as people and material, traffic flow is of great significance for understanding the characteristics of spatial connection. In this paper, Harbin-Changchun agglomeration proposed by China's New Urbanization Plan (2014-2020) is taken as a research object. With the data of intercity passenger traffic flow including highway and railway passenger trips between 73 county-level spatial units in the research area, a traffic flow model is constructed to measure the intensity of spatial connection. -

The Construction and Optimization of Ecological Security Pattern in the Harbin-Changchun Urban Agglomeration, China
Article The Construction and Optimization of Ecological Security Pattern in the Harbin-Changchun Urban Agglomeration, China Rong Guo 1, Tong Wu 1,*, Mengran Liu 2, Mengshi Huang 1, Luigi Stendardo 3 and Yutong Zhang 4 1 Key Laboratory of Cold Region Urban and Rural Human Settlement Environment Science and Technology, Ministry of Industry and Information Technology. School of Architecture, Harbin Institute of Technology, Harbin 150006, China; [email protected] (R.G.); [email protected] (M.H.) 2 Key Laboratory of Forest Plan Ecology, Ministry of Education, Northeast Forestry University, Harbin 150040, China; [email protected] 3 Department of Civil, Architectural and Environmental Engineering, Padova University, 35131 Padova, Italy; [email protected] 4 Department of Agriculture and Forestry Economic Management, School of Economic Management, Northeast Forestry University, Harbin 150040, China; [email protected] * Correspondence: [email protected]; Tel.: +86-135-0360-9537 Received: 03 March 2019; Accepted: 30 March 2019; Published: 2 April 2019 Abstract: Urban agglomerations have become a new geographical unit in China, breaking the administrative fortresses between cities, which means that the population and economic activities between cities will become more intensive in the future. Constructing and optimizing the ecological security pattern of urban agglomerations is important for promoting harmonious social-economic development and ecological protection. Using the Harbin-Changchun urban agglomeration as a case study, we have identified ecological sources based on the evaluation of ecosystem functions. Based on the resistance surface modified by nighttime light (NTL) data, the potential ecological corridors were identified using the least-cost path method, and key ecological corridors were extracted using the gravity model. -

Two-Dimensional Water Environment Numerical Simulation Research Based on EFDC in Mudan River, Northeast China
2015 IEEE European Modelling Symposium Two-Dimensional Water Environment Numerical Simulation Research Based on EFDC in Mudan River,Northeast China Gula Tang1, 2, 3*, Jing Li4, Yunqiang Zhu1, Zhaoliang Li2, Françoise Nerry2 1 State Key Laboratory of Resources and Environmental Information Systems, Institute of Geographical Sciences and Natural Resources Research, Chinese Academy of Sciences, Beijing, China 2 ICube (UMR 7357), UdS,CNRS, Strasbourg, France 3 University of Chinese Academy of Sciences, Beijing, China 4 Institute of Environmental Sciences of Heilongjiang Province, Heilongjiang, China e-mail:[email protected] Abstract—This paper establishes a two-dimensional (2D) from which massive industrial effluents and sanitary numerical simulation model for water environment of sewage are discharged into the Mudan River in various Mudan River using hydrodynamic and water quality model means, bringing low water quality in Mudan River. based on EFDC. It simulates the migration of CODCr and According to findings by Yu HAO et al. [1], water bodies in NH3-N in urban sections of the trunk stream in glacial and urban sections of Mudan River trunk stream are in organic non-glacial periods. It also calibrates and verifies bed pollution, with major over standard factors of roughness and integrated attenuation coefficient of pollutants. As the findings reveal, bed roughness and permanganate and ammonia nitrogen. integrated attenuation coefficient of pollutants vary Water quality modeling plays an increasingly significantly in icebound and non-icebound seasons: important role in water environment protection, roughness coefficient in icebound season is higher than that management and decision-making support. It helps to in non-icebound season while attenuation rate in icebound predict and assess water quality responses to natural season is lower than that in non-icebound season. -

Study on GIS Visualization in Evaluation of the Human Living Environment in Shenyang-Dalian Urban Agglomeration
Hindawi Publishing Corporation Scientifica Volume 2016, Article ID 7462832, 10 pages http://dx.doi.org/10.1155/2016/7462832 Research Article Study on GIS Visualization in Evaluation of the Human Living Environment in Shenyang-Dalian Urban Agglomeration Kang Hou, Jieting Zhou, Xuxiang Li, and Shengbin Ge School of Human Settlements and Civil Engineering, Xi’an Jiaotong University, Xi’an 710049, China Correspondence should be addressed to Xuxiang Li; [email protected] Received 7 December 2015; Revised 1 March 2016; Accepted 31 March 2016 Academic Editor: Francisco Ayuga Copyright © 2016 Kang Hou et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Analysis of human living environmental quality of Shenyang-Dalian urban agglomerations has important theoretical and practical significance in rapid development region. A lot of investigations have been carried for Shenyang-Dalian urban agglomerations, including 38 counties. Based on the carrying capacity of resources, natural and socioeconomic environmental factors and regional changes of human living environmental evaluation are analyzed with the application of geographic information systems (GIS) software. By using principal component analysis (PCA) model and natural breaks classification (NBC) method, the evaluation results are divided into five categories. The results show thatuman theh living environmental evaluation (HLEE) indexes of Dalian, Shenyang, and Liaoyang are higher than other counties. Among these counties, the human living environmental evaluation (HLEE) indexes of coastal counties are significantly higher than inland counties. The range of the human living environmental evaluation index in most of the study area is at III, IV,and V levels, accounting for 80.01%. -
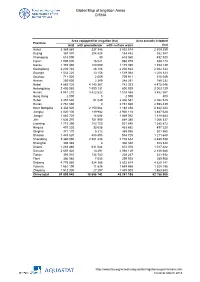
Global Map of Irrigation Areas CHINA
Global Map of Irrigation Areas CHINA Area equipped for irrigation (ha) Area actually irrigated Province total with groundwater with surface water (ha) Anhui 3 369 860 337 346 3 032 514 2 309 259 Beijing 367 870 204 428 163 442 352 387 Chongqing 618 090 30 618 060 432 520 Fujian 1 005 000 16 021 988 979 938 174 Gansu 1 355 480 180 090 1 175 390 1 153 139 Guangdong 2 230 740 28 106 2 202 634 2 042 344 Guangxi 1 532 220 13 156 1 519 064 1 208 323 Guizhou 711 920 2 009 709 911 515 049 Hainan 250 600 2 349 248 251 189 232 Hebei 4 885 720 4 143 367 742 353 4 475 046 Heilongjiang 2 400 060 1 599 131 800 929 2 003 129 Henan 4 941 210 3 422 622 1 518 588 3 862 567 Hong Kong 2 000 0 2 000 800 Hubei 2 457 630 51 049 2 406 581 2 082 525 Hunan 2 761 660 0 2 761 660 2 598 439 Inner Mongolia 3 332 520 2 150 064 1 182 456 2 842 223 Jiangsu 4 020 100 119 982 3 900 118 3 487 628 Jiangxi 1 883 720 14 688 1 869 032 1 818 684 Jilin 1 636 370 751 990 884 380 1 066 337 Liaoning 1 715 390 783 750 931 640 1 385 872 Ningxia 497 220 33 538 463 682 497 220 Qinghai 371 170 5 212 365 958 301 560 Shaanxi 1 443 620 488 895 954 725 1 211 648 Shandong 5 360 090 2 581 448 2 778 642 4 485 538 Shanghai 308 340 0 308 340 308 340 Shanxi 1 283 460 611 084 672 376 1 017 422 Sichuan 2 607 420 13 291 2 594 129 2 140 680 Tianjin 393 010 134 743 258 267 321 932 Tibet 306 980 7 055 299 925 289 908 Xinjiang 4 776 980 924 366 3 852 614 4 629 141 Yunnan 1 561 190 11 635 1 549 555 1 328 186 Zhejiang 1 512 300 27 297 1 485 003 1 463 653 China total 61 899 940 18 658 742 43 241 198 52