University of Groningen Genome Sequencing and Analysis Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Annual Conference Abstracts
ANNUAL CONFERENCE 14-17 April 2014 Arena and Convention Centre, Liverpool ABSTRACTS SGM ANNUAL CONFERENCE APRIL 2014 ABSTRACTS (LI00Mo1210) – SGM Prize Medal Lecture (LI00Tu1210) – Marjory Stephenson Climate Change, Oceans, and Infectious Disease Prize Lecture Dr. Rita R. Colwell Understanding the basis of antibiotic resistance University of Maryland, College Park, MD, USA as a platform for early drug discovery During the mid-1980s, satellite sensors were developed to monitor Laura JV Piddock land and oceans for purposes of understanding climate, weather, School of Immunity & Infection and Institute of Microbiology and and vegetation distribution and seasonal variations. Subsequently Infection, University of Birmingham, UK inter-relationships of the environment and infectious diseases Antibiotic resistant bacteria are one of the greatest threats to human were investigated, both qualitatively and quantitatively, with health. Resistance can be mediated by numerous mechanisms documentation of the seasonality of diseases, notably malaria including mutations conferring changes to the genes encoding the and cholera by epidemiologists. The new research revealed a very target proteins as well as RND efflux pumps, which confer innate close interaction of the environment and many other infectious multi-drug resistance (MDR) to bacteria. The production of efflux diseases. With satellite sensors, these relationships were pumps can be increased, usually due to mutations in regulatory quantified and comparatively analyzed. More recent studies of genes, and this confers resistance to antibiotics that are often used epidemic diseases have provided models, both retrospective and to treat infections by Gram negative bacteria. RND MDR efflux prospective, for understanding and predicting disease epidemics, systems not only confer antibiotic resistance, but altered expression notably vector borne diseases. -

Rehmannia Glutinosa-Monocultured Rhizosphere Soil
Comparative Metaproteomic Analysis on Consecutively Rehmannia glutinosa-Monocultured Rhizosphere Soil Linkun Wu1,2, Haibin Wang1,2., Zhixing Zhang1,2., Rui Lin2,3, Zhongyi Zhang1,4, Wenxiong Lin1,2* 1 School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 2 Agroecological Institute, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 3 College of Oceanography and Environmental Science, Xiamen University, Xiamen, Fujian, China, 4 Institute of Chinese Medicinal Materials, Henan Agriculture University, Zhengzhou, Henan, China Abstract Background: The consecutive monoculture for most of medicinal plants, such as Rehmannia glutinosa, results in a significant reduction in the yield and quality. There is an urgent need to study for the sustainable development of Chinese herbaceous medicine. Methodology/Principal Findings: Comparative metaproteomics of rhizosphere soil was developed and used to analyze the underlying mechanism of the consecutive monoculture problems of R. glutinosa. The 2D-gel patterns of protein spots for the soil samples showed a strong matrix dependency. Among the spots, 103 spots with high resolution and repeatability were randomly selected and successfully identified by MALDI TOF-TOF MS for a rhizosphere soil metaproteomic profile analysis. These proteins originating from plants and microorganisms play important roles in nutrient cycles and energy flow in rhizospheric soil ecosystem. They function in protein, nucleotide and secondary metabolisms, signal transduction and resistance. Comparative metaproteomics analysis revealed 33 differentially expressed protein spots in rhizosphere soil in response to increasing years of monoculture. Among them, plant proteins related to carbon and nitrogen metabolism and stress response, were mostly up-regulated except a down-regulated protein (glutathione S-transferase) involving detoxification. -
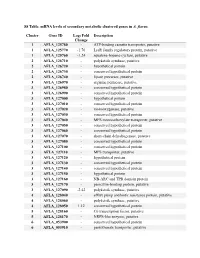
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Supporting Information
Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were plated on YPD with or without 50 mM HU and incubated at 30°C or 37°C for 3 days. Figure S2. Localization of Arp6 and Swr1 on chromosome 3. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) are compared. The position of Tel 3L, Tel 3R, CEN3, and the RP gene are shown under the panels. Figure S3. Localization of Arp6 and Swr1 on chromosome 4. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) in the whole chromosome region are compared. The position of Tel 4L, Tel 4R, CEN4, SWR1, and RP genes are shown under the panels. Figure S4. Localization of Arp6 and Swr1 on the region including the SWR1 gene of chromosome 4. The binding of Arp6- FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) are compared. The position and orientation of the SWR1 gene is shown. Figure S5. Localization of Arp6 and Swr1 on chromosome 5. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) are compared. The position of Tel 5L, Tel 5R, CEN5, and the RP genes are shown under the panels. Figure S6. Preferential localization of Arp6 and Swr1 in the 5′ end of genes. Vertical bars represent the binding ratio of proteins in each locus. -

Genes in Eyecare Geneseyedoc 3 W.M
Genes in Eyecare geneseyedoc 3 W.M. Lyle and T.D. Williams 15 Mar 04 This information has been gathered from several sources; however, the principal source is V. A. McKusick’s Mendelian Inheritance in Man on CD-ROM. Baltimore, Johns Hopkins University Press, 1998. Other sources include McKusick’s, Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders. Baltimore. Johns Hopkins University Press 1998 (12th edition). http://www.ncbi.nlm.nih.gov/Omim See also S.P.Daiger, L.S. Sullivan, and B.J.F. Rossiter Ret Net http://www.sph.uth.tmc.edu/Retnet disease.htm/. Also E.I. Traboulsi’s, Genetic Diseases of the Eye, New York, Oxford University Press, 1998. And Genetics in Primary Eyecare and Clinical Medicine by M.R. Seashore and R.S.Wappner, Appleton and Lange 1996. M. Ridley’s book Genome published in 2000 by Perennial provides additional information. Ridley estimates that we have 60,000 to 80,000 genes. See also R.M. Henig’s book The Monk in the Garden: The Lost and Found Genius of Gregor Mendel, published by Houghton Mifflin in 2001 which tells about the Father of Genetics. The 3rd edition of F. H. Roy’s book Ocular Syndromes and Systemic Diseases published by Lippincott Williams & Wilkins in 2002 facilitates differential diagnosis. Additional information is provided in D. Pavan-Langston’s Manual of Ocular Diagnosis and Therapy (5th edition) published by Lippincott Williams & Wilkins in 2002. M.A. Foote wrote Basic Human Genetics for Medical Writers in the AMWA Journal 2002;17:7-17. A compilation such as this might suggest that one gene = one disease. -

2016 Joint Meeting Program
April 15 – 17, 2016 Fairmont Chicago Millennium Park • Chicago, Illinois The AAP/ASCI/APSA conference is jointly provided by Boston University School of Medicine and AAP/ASCI/APSA. Meeting Program and Abstracts www.jointmeeting.org www.jointmeeting.org Special Events at the 2016 AAP/ASCI/APSA Joint Meeting Friday, April 15 Saturday, April 16 ASCI President’s Reception ASCI Food and Science Evening 6:15 – 7:15 p.m. 6:30 – 9:00 p.m. Gold Room The Mid-America Club, Aon Center ASCI Dinner & New Member AAP Member Banquet Induction Ceremony (Ticketed guests only) (Ticketed guests only) 7:00 – 10:00 p.m. 7:30 – 9:45 p.m. Imperial Ballroom, Level B2 Rouge, Lobby Level How to Solve a Scientific Puzzle: Speaker: Clara D. Bloomfield, MD Clues from Stockholm and Broadway The Ohio State University Comprehensive Cancer Center Speaker: Joe Goldstein, MD APSA Welcome Reception & University of Texas Southwestern Medical Center at Dallas Presidential Address APSA Dinner (Ticketed guests only) 9:00 p.m. – Midnight Signature Room, 360 Chicago, 7:30 – 9:00 p.m. John Hancock Center (off-site) Rouge, Lobby Level Speaker: Daniel DelloStritto, APSA President Finding One’s Scientific Niche: Musings from a Clinical Neuroscientist Speaker: Helen Mayberg, MD, Emory University Dessert Reception (open to all attendees) 10:00 p.m. – Midnight Imperial Foyer, Level B2 Sunday, April 17 APSA Future of Medicine and www.jointmeeting.org Residency Luncheon Noon – 2:00 p.m. Rouge, Lobby Level 2 www.jointmeeting.org Program Contents General Program Information 4 Continuing Medical Education Information 5 Faculty and Speaker Disclosures 7 Scientific Program Schedule 9 Speaker Biographies 16 Call for Nominations: 2017 Harrington Prize for Innovation in Medicine 26 AAP/ASCI/APSA Joint Meeting Faculty 27 Award Recipients 29 Call for Nominations: 2017 Harrington Scholar-Innovator Award 31 Call for Nominations: George M. -

Electric Supporting Information (ESI) Crystal Structure and Functional
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2018 Electric Supporting Information (ESI) Crystal structure and functional analysis of large-terpene synthase belonging to a newly found subclass Masahiro Fujihashi,a* Tsutomu Sato,b* Yuma Tanaka,a Daisuke Yamamoto,a Tomoyuki Nishi,b Daijiro Ueda,b Mizuki Murakami,b Yoko Yasuno,c Ai Sekihara,c Kazuma Fuku,c Tetsuro Shinadac & Kunio Mikia* a. Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: [email protected], [email protected] b. Department of Applied Biological Chemistry, Faculty of Agriculture, and Graduate School of Science and Technology, Niigata University, 8050 Ikarashi-2, Niigata 950-2181, Japan. E-mail: [email protected] c. Graduate School of Science, Osaka City University, 3-3-138 Sugimoto, Sumiyoshi, Osaka 558-8585, Japan. S1 Table of Contents Experimental Procedures S3 General procedure S3 Vector construction, expression and purification of BalTS S3 Crystallization and crystallographic analysis of balTS S3 Oligomeric state analysis S4 Analysis of the dimer geometry S4 Vector construction, expression and purification of BsuTS S4 Homology modeling of BsuTS S4 Enzymatic assays S5 Isolation and identification of β-springen S5 Movie Captions S6 Author Contributions S6 Figures Fig. S1 S7 Fig. S2 S8 Fig. S3 S9 Fig. S4 S10 Fig. S5 S17 Fig. S6 S18 Fig. S7, S8 S19 Fig. S9 S20 Fig. S10 S21 Fig. S11 S22 Tables Table S1 S23 Table S2 S24 References S27 S2 Experimental Procedures General procedure NMR spectra were recorded using a Bruker DPX 400 spectrometer at 400 MHz for proton (1H) and 100 MHz for carbon (13C). -

Teleomorph Hypocrea Jecorina)
Downloaded from orbit.dtu.dk on: Dec 20, 2017 Unraveling the Secondary Metabolism of the Biotechnological Important Filamentous Fungus Trichoderma reesei ( Teleomorph Hypocrea jecorina) Jørgensen, Mikael Skaanning; Larsen, Thomas Ostenfeld; Mortensen, Uffe Hasbro; Aubert, Dominique Publication date: 2013 Document Version Publisher's PDF, also known as Version of record Link back to DTU Orbit Citation (APA): Jørgensen, M. S., Larsen, T. O., Mortensen, U. H., & Aubert, D. (2013). Unraveling the Secondary Metabolism of the Biotechnological Important Filamentous Fungus Trichoderma reesei ( Teleomorph Hypocrea jecorina). Kgs. Lyngby: Technical University of Denmark (DTU). General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Unraveling the Secondary Metabolism of the Biotechnological Important Filamentous Fungus Trichoderma reesei (Teleomorph Hypocrea jecorina) MJ-T-030 Mikael Skaanning Jørgensen Ph.D. Thesis November 2012 MJ-T-020 MJ-T-032 28 8 20 27 5 28 1 30 17 8 24 25 9 25 31 13 15 26 16 4 24 12 32 18 11 3 14 13 19 10 5 7 2 9 2521 22 23 Unraveling the Secondary Metabolism of the Biotechnological Important Filamentous Fungus Trichoderma reesei (Teleomorph Hypocrea jecorina) Mikael Skaanning Jørgensen Ph.D. -

George Michael Humphrey Birchenough
GEORGE MICHAEL HUMPHREY BIRCHENOUGH Analysis of intestinal factors contributing to the age- dependency of systemic neuropathogenic Escherichia coli K1 infection in the neonatal rat Thesis submitted in accordance with the requirements of the UCL School of Pharmacy for the degree of Doctor of Philosophy Microbiology Group, Department of Pharmaceutics, UCL School of Pharmacy July 2012 PLAGIARISM STATEMENT This thesis describes research conducted in the UCL School of Pharmacy between October 2008 and July 2012 under the supervision of Professor Peter W. Taylor. I certify that the research described is original and that any parts of the work that have been conducted by collaboration are clearly indicated. I also certify that I have written all the text herein and have clearly indicated by suitable citation any part of the dissertation that has already appeared in publication. Signature: Date: Acknowledgements Firstly I wish to thank my supervisor, Professor Peter Taylor, for giving me the opportunity to work on such an interesting and rewarding project. Your continued support and enthusiasm has been a constant source of encouragement and I greatly appreciate all the advice and help (both scientific and general!) that you have provided over the last four years. I owe you a lot of beer. I also wish to thank my amazing parents for all their love and support over the eight years of my higher education. Without your enthusiasm and belief I would not have been able to follow this path. I sincerely promise I will now get a job! Furthermore, I wish to thank colleagues at the London School of Hygiene & Tropical Medicine, Dr. -

The Inducers 1,3-Diaminopropane and Spermidine Cause The
JOURNAL OF PROTEOMICS 85 (2013) 129– 159 Available online at www.sciencedirect.com www.elsevier.com/locate/jprot The inducers 1,3-diaminopropane and spermidine cause the reprogramming of metabolism in Penicillium chrysogenum, leading to multiple vesicles and penicillin overproduction Carlos García-Estradaa,⁎, Carlos Barreiroa, Mohammad-Saeid Jamia, Jorge Martín-Gonzáleza, Juan-Francisco Martínb,⁎⁎ aINBIOTEC, Instituto de Biotecnología de León, Avda. Real no. 1, Parque Científico de León, 24006 León, Spain bÁrea de Microbiología, Departamento de Biología Molecular, Universidad de León, Campus de Vegazana s/n; 24071 León, Spain ARTICLE INFO ABSTRACT Article history: In this article we studied the differential protein abundance of Penicillium chrysogenum in Received 21 February 2013 response to either 1,3-diaminopropane (1,3-DAP) or spermidine, which behave as inducers Accepted 15 April 2013 of the penicillin production process. Proteins were resolved in 2-DE gels and identified by Available online 30 April 2013 tandem MS spectrometry. Both inducers produced largely identical changes in the proteome, suggesting that they may be interconverted and act by the same mechanism. Keywords: The addition of either 1,3-DAP or spermidine led to the overrepresentation of the last 1,3-diaminopropane enzyme of the penicillin pathway, isopenicillin N acyltransferase (IAT). A modified form of Spermidine the IAT protein was newly detected in the polyamine-supplemented cultures. Both Penicillin inducers produced a rearrangement of the proteome resulting in an overrepresentation of Penicillium chrysogenum enzymes involved in the biosynthesis of valine and other precursors (e.g. coenzyme A) of Vesicles penicillin. Interestingly, two enzymes of the homogentisate pathway involved in the degradation of phenylacetic acid (a well-known precursor of benzylpenicillin) were reduced following the addition of either of these two inducers, allowing an increase of the phenylacetic acid availability. -

Nucleosome Assembly and Histone H3 Methylation During Dna Replication
NUCLEOSOME ASSEMBLY AND HISTONE H3 METHYLATION DURING DNA REPLICATION TxrwTom Rolef Ben-Shahar A thesis submitted for the degree of Ph.D. at The University of London July, 2004 Cancer Research UK London Research Institute, Clare Hall Laboratories, South Mimms, Hertfordshire, EN6 3LD and University College London, Gower Street, London, WC1 6BT ProQuest Number: 10014844 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest. ProQuest 10014844 Published by ProQuest LLC(2016). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code. Microform Edition © ProQuest LLC. ProQuest LLC 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106-1346 ACKNOWLEDGEMENTS To begin with, I would like to acknowledge my mistakes, scientific and others. Hopefully the future will show that I have learned what I could from them. My work has been funded by Cancer Research UK and with a B’NAI BRITH scholarship, and for that I am grateful. I would like to thank Alain Verreault for his supervision, for our scientific conversations and for allowing me to make my own mistakes. My gratitude is given to Munah Abdul-Rauf for being a colleague, collaborator, friend and sister; to Michal Goldberg, Niki Hawks and to other friends and colleagues in Clare-Hall. -

Structural Basis for the Activation of Phenylalanine in the Non-Ribosomal Biosynthesis of Gramicidin S
The EMBO Journal Vol.16 No.14 pp.4174–4183, 1997 Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S Elena Conti1,2, Torsten Stachelhaus3, the peptide product. Each amino acid is activated by Mohamed A.Marahiel3 and Peter Brick1,4 adenylation of its carboxylate group with ATP and then transferred to the thiol group of an enzyme-bound phospho- 1Biophysics Section, Blackett Laboratory, Imperial College, pantetheine cofactor for possible modification and the 3 London SW7 2BZ, UK and Biochemie/Fachbereich Chemie, elongation reaction (Stachelhaus and Marahiel, 1995a; Philipps-Universita¨t Marburg, D-35032 Marburg, Germany Kleinkauf and von Do¨hren, 1996). 2 Present address: Laboratory of Molecular Biophysics, The cloning and sequencing of several peptide synthe- Rockefeller University, New York, NY10021, USA tase genes have revealed a conserved and ordered modular 4Corresponding author organization. Each module encodes a functional building unit containing ~1000 amino acids, which specifically The non-ribosomal synthesis of the cyclic peptide recognizes a single amino acid. Within such a protein antibiotic gramicidin S is accomplished by two large template-directed peptide biosynthesis, the occurrence and multifunctional enzymes, the peptide synthetases 1 and specific order of the modules in the genomic DNA dictate 2. The enzyme complex contains five conserved subunits the number and sequence of the amino acids to be of ~60 kDa which carry out ATP-dependent activation incorporated into the resulting oligopeptide. The modular of specific amino acids and share extensive regions of arrangement of peptide synthetases closely parallels the sequence similarity with adenylating enzymes such multienzyme complexes responsible for the biogenesis of as firefly luciferases and acyl-CoA ligases.